This GUI and associated scripts accompany the paper "Brain-wide Organization of Neuronal Activity and Convergent Sensorimotor Transformations in Larval Zebrafish" (Neuron 2018, also see preprint on biorxiv). The GUI is developed to interactively explore and visualize functional imaging data. The data can be curated in the GUI quickly in terms of selecting a subset of cells or time-points. Different analyses can be performed in quick succession on the exact data subset that is visualized. This platform vastly simplifies bookkeeping and visualizations, and was developed for prototyping many custom analyses that operate on single-animal data. The associated scripts extend the same analysis framework to analyses across groups of animals.
All data curated in the GUI can also be easily edited in the MATLAB workspace (see demo\demo_workspace_interactive.m) or in script form (see demo\demo_script.m); import back to the GUI for further exploration and visualization.
The data consists of recordings of whole-brain recordings from larval zebrafish, fictively behaving in a virtual environment, imaged using a light-sheet microscope. The hardware system is detailed in Vladimirov, Mu, Kawashima, Bennett, Yang, Looger, Keller, Freeman and Ahrens, Nature Methods, 2014 (https://www.nature.com/articles/nmeth.3040).
The data were collected by Yu Mu (muy (at) janelia.hhmi.org) in the Ahrens lab.
The data is provided openly for academic, non-commercial use under a Creative Commons license (that can be downloaded here). Use of the data should be accompanied by a citation to the source paper,
Brain-wide Organization of Neuronal Activity and Convergent Sensorimotor Transformations in Larval Zebrafish.
Xiuye Chen*, Yu Mu*, Yu Hu*, Aaron T. Kuan*, Maxim Nikitchenko, Owen Randlett, Alex B. Chen, Jeffery P. Gavornik, Haim Sompolinsky, Florian Engert, and Misha B. Ahrens (*: equal contributions)
Neuron, 2018
DOI: https://doi.org/10.1016/j.neuron.2018.09.042
The data is available for download on FigShare: https://doi.org/10.25378/janelia.7272617
Questions about the data or code should be directed to xiuye.chen (at) gmail and ahrensm (at) janelia.hhmi.org.
While the code here is provided "as is" without guarantee, we are always happy to communicate about the data and any findings based on it, please get in touch.
-
Download the data.
-
To point the code to the data folder, download the code, and find the sub-folder "dir setup". Update directory in "GetCurrentDataDir.m" to the folder where the downloaded data is now stored. Update directory in "GetOutputDataDir.m" to the same directory as well (you can change this later as long as outputDir contains "VAR_new.mat"). ("GetFishDirectories.m" and "GetNestedDataDir.m" are for pre-processing steps, not used here.)
-
Run "LoadGUI.m" to open GUI. (take note to add full code dir to MATLAB search path)
-
Load data from a fish by selecting the fish ID number in the dropdown menu "Load fish #" under "General" tab. (If you downloaded data for only 1 example fish, load fish 6.)
-
Main code is in "GUI_FishExplorer.m". Follow tips at the top of the code to navigate.
-
To see some example selections of cells, go to tab "Saved Clusters", and browse drop-down menus "Cluster" and "ClusterGroup" (which is groups of "Clusters").
-
In tab "Operations", use the "Back" and "Forward" button to access the previous views. "Choose cluster range" takes inputs like "1-3,5" or "2:end". Cluster range are indicated by the numbers next to the color bar.
(For more details on this gif, see FishExplorer\demo\FuncGUI_with_ZBrain_demo.pdf)
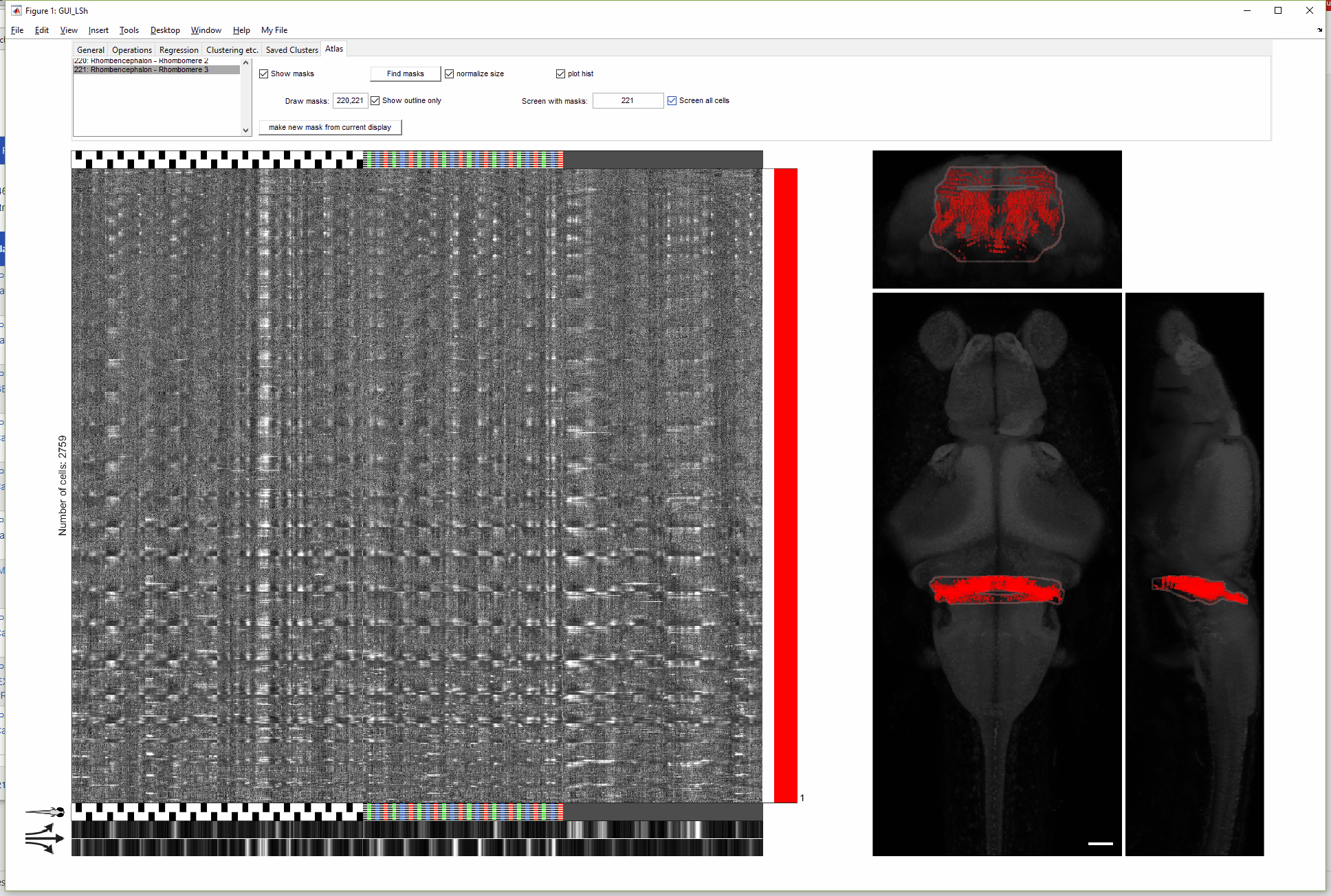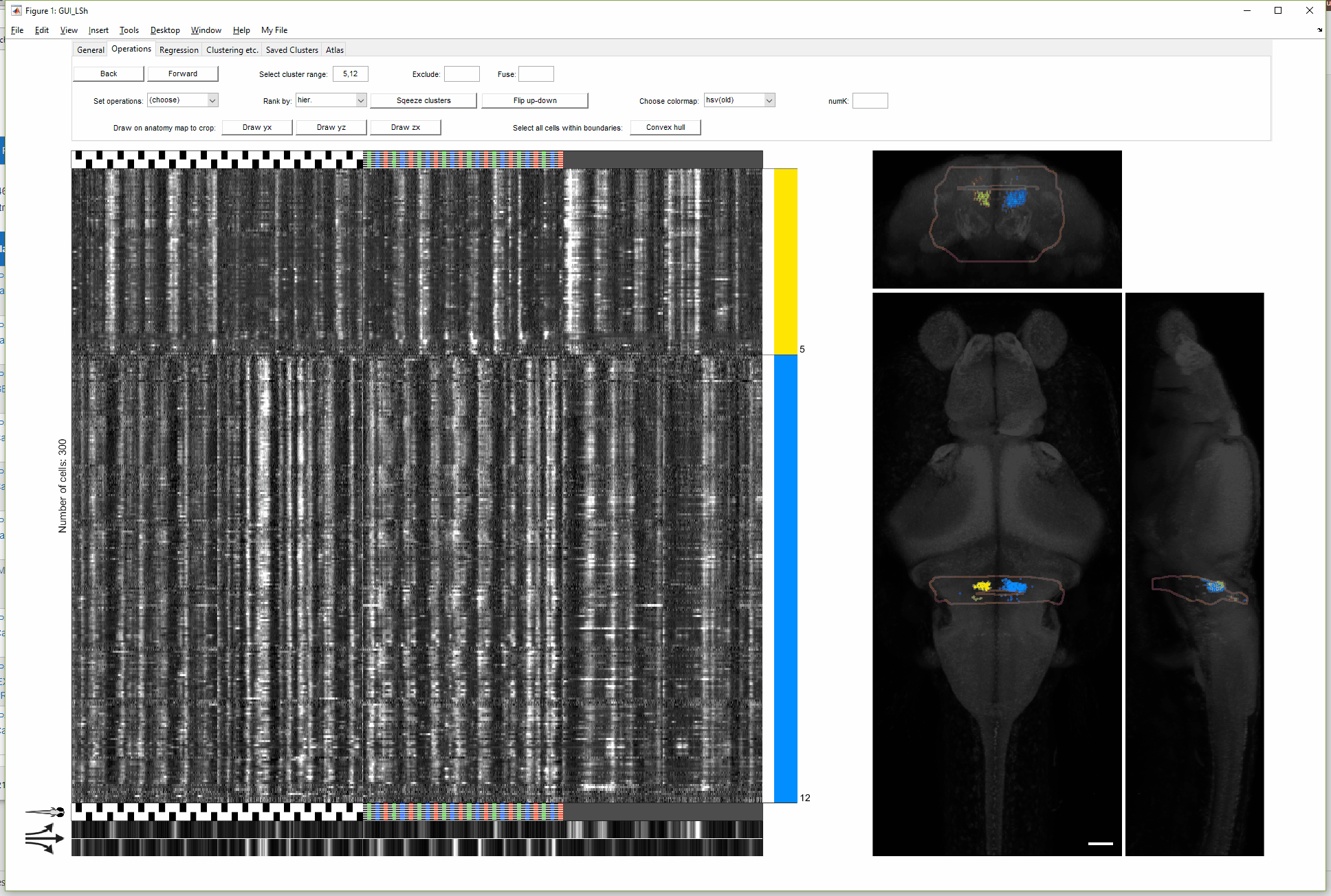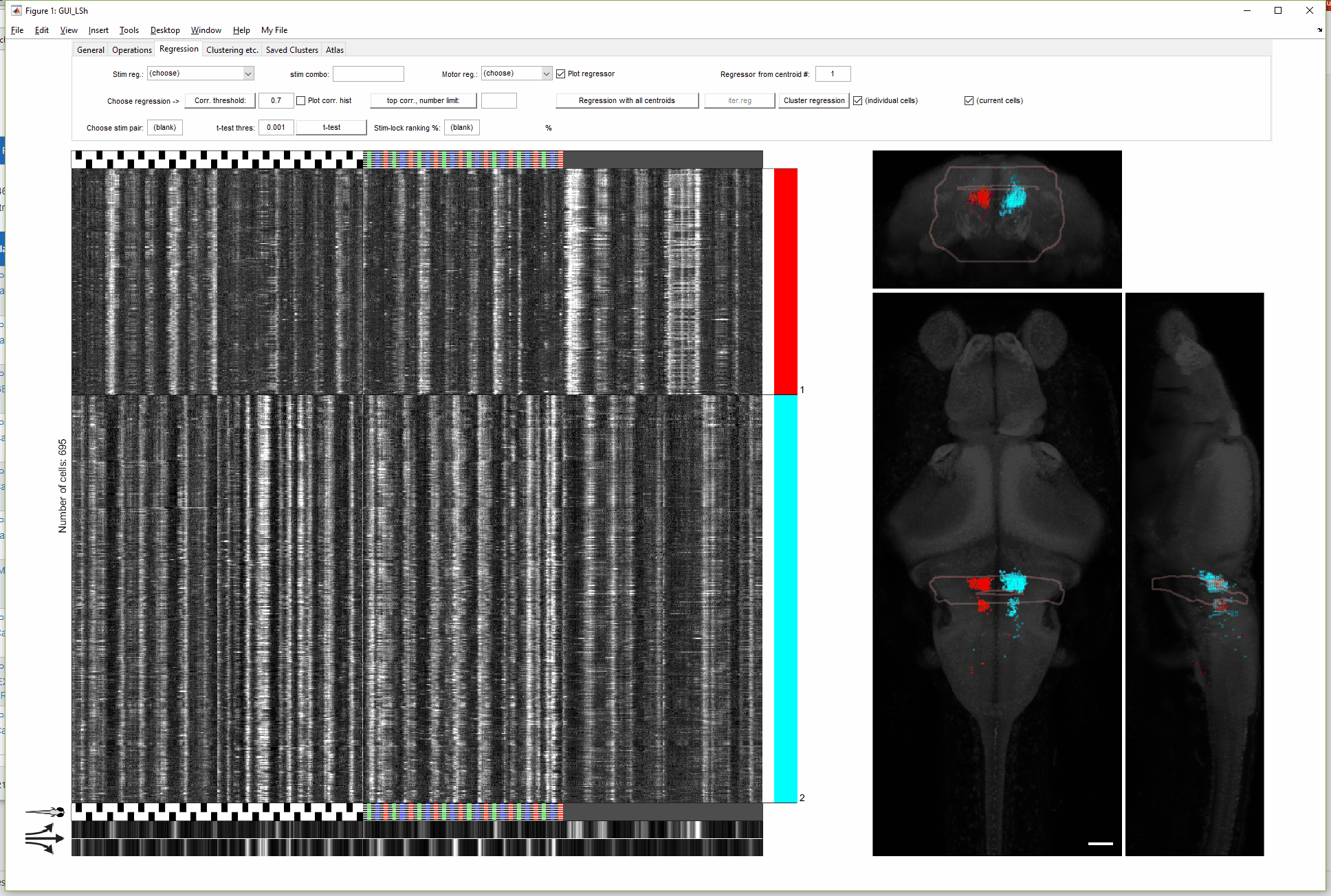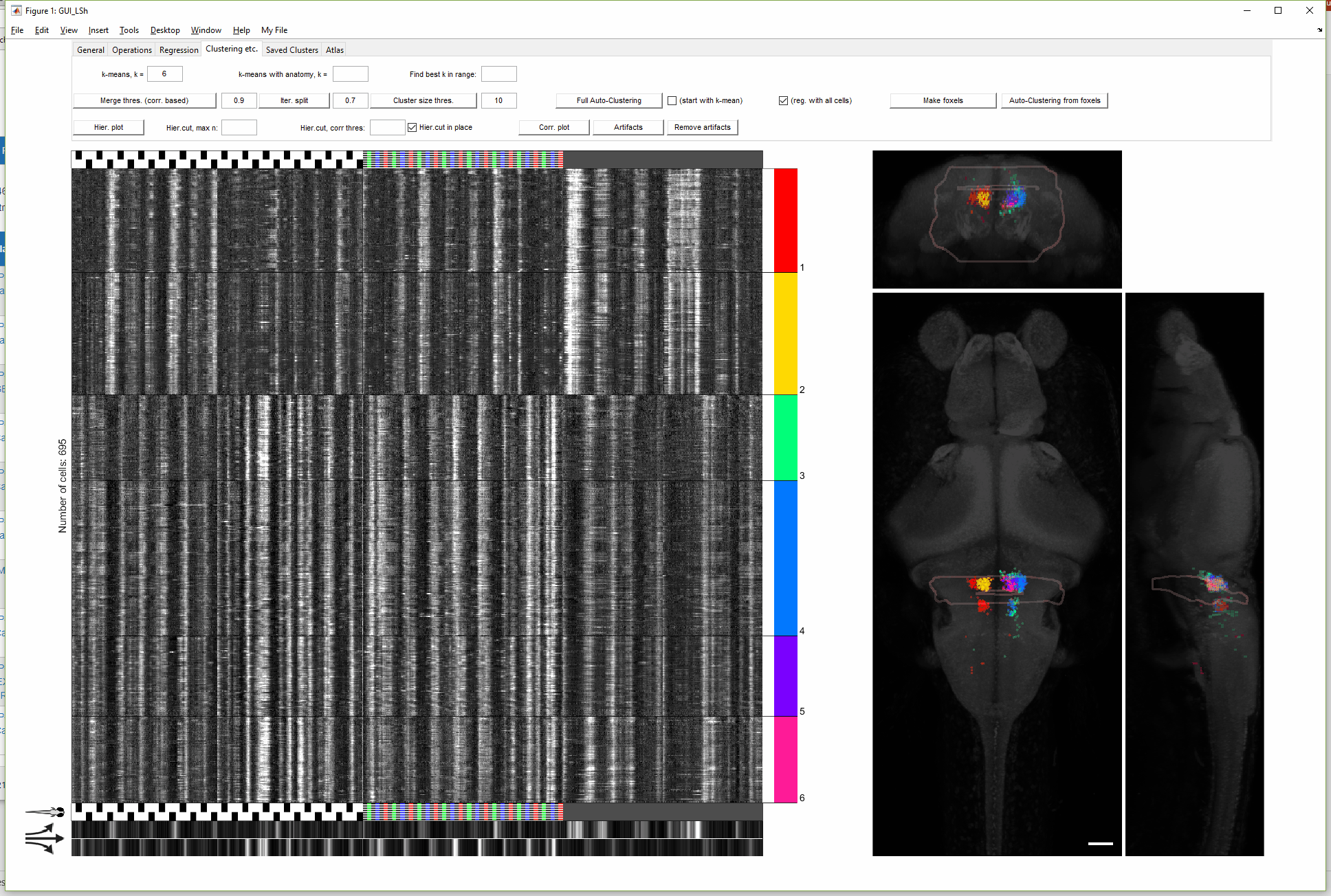
A newer version of this GUI (with more limited functionality) can be found here, developed to conveniently take the output of a popular preprocessing package, Suite2p, as input data.
Questions about the data or code should be directed to xiuye.chen (at) gmail and ahrensm (at) janelia.hhmi.org.