New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Reducing refractory period violation for tetrode data #311
Comments
|
What version of the software are you using? If you want a quick feedback, the best would be to share your data. Is it something possible for you ? But so far, what your did with the parameters seems legit. Except that in recent version, the dispersion parameter is not used anymore. |
|
Thank you for the quick reply. I'm using version 1.0.1. |
|
I had a look to your data, and the .params you sent me. First, N_t=1 is way too small, and we can see your templates are cropped. Cropped templates means non optimal clustering/fitting. So bring back the value to 2 or 3. Secondly, to better understand what is done by the software, set make_plots = png in the [clustering] section. It will generate plots of the local clustering, in the results folder/plot. Using the master branch, everything looks rather fine to me, so which version of the software are you using? Please update to master, such that we are sure to be in sync From the debug plots, we can see that the clustering are performing rather decently, with no "obvious" errors. If you want to get more clusters, I would not play with sensitivity, but rather with the merging_param value. Since you have 16 local merges, it will reduce this number, and you'll have more clusters (at the cost of a higher overclustering maybe). So set 1 or even less. Regarding the final templates, you do have some without RPV, and some with. This is also because quite a lot are defined on only single channel. But to me, I do not see the behavior that you are showing |
|
That sounds little bit promising. I now use the latest master branch, and try with the default parameters and changing Here is my .prams file Thank you for your time. |
|
Decreasing the spike_thresh to 4.25 to very low value does not mean that you'll get more spikes, and that you'll get the clusters you had with a higher spike_thresh (at 6) plus some new ones. This might be counter intuitive, but let me explain. For the clustering, we select 10000 spikes per electrodes to cluster from. If you have a too low spike_thresh, these selected spikes might be just noise, and the large spikes might be under-represented. We have ways to somehow try to compensate for that, but this is not perfect. This is why I would not really advise to decrease spike_thresh too much, except if your data have a very high SNR. I'll look again at the data for the RPV |
|
I have tried several other recordings but all clusters had RPV issue. Could you possibly share your parameter file info that worked well? Thank you very much. |
|
Use master branch (1.0.2) and the attached parameter file (renamed as .txt) |
|
Thanks a lot for the parameter file. It resulted in an error during 'estimating the templates'. Sorry if I am making stupid mistake. |
|
Yes, I noticed the bug, this has been patched. Sorry for that. Please upgrade to master, and I'll make a release asap |
|
Thanks for the update. In my environment, all resulting clusters are still having RPV... I am not sure what makes it wrong in my environment. I attach the log file. The log file is not full as it had tens of thousands of lines with DEBUG [matplotlib.font_manager] and DEBUG [matplotlib.ticker], which I assume are not relevant here, and I deleted them manually (but why are there so many?). I may have accidentally deleted lines with DEBUG [circus.clustering]. Thanks a lot for your time. |
|
If you are a windows user, a silent bug recently spotted in windows only might be the reason for the problem... Sorry for that, there was an int overflow not triggering errors. Try upgrading to master, and hopefully it will be better |
|
Thank you for the update. Meanwhile, I tried more systematic testing of the parameters i.e. changing one parameter at once, occasionally to an extreme value.
Changing other parameters alone did not give obvious improvement. Is it feasible and does it make sense to have an option to assign spikes in the refractory period to separate clusters? |
|
But are you using windows ot not? Because as said, it you are, then there was a bug in the amplitudes that could lead to noise contamination... |
|
Yes, I am using Windows. I will try with the latest commit again. Thank you. |
|
In the master branch, yes. Amplitudes should be much better (with default parameter, i.e. fine_ampltiude = True) |
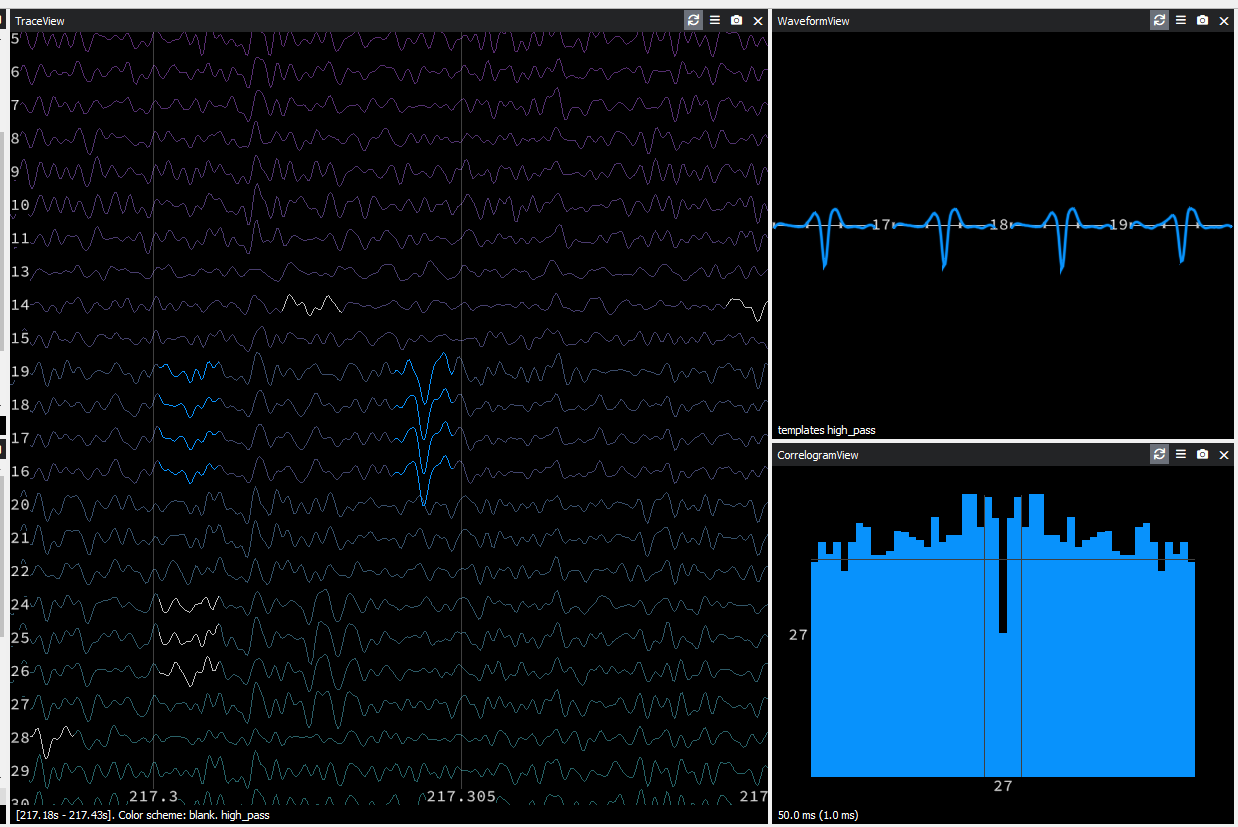
I am new to SpyKING CIRCUS and I much appreciate the software. I do have a problem on sorting quality.

I have 8-tetrode / 32-channel recordings and raw data are already band pass filtered for spike analysis (1-6 kHz). Some channels have good s/n ratio. This is example raw traces from a tetrode.
These traces are ones with the best quality I can get in my recording setup and I am testing the algorithm with these data.
With the default setting, I got relatively few clusters (<3 clusters per channel) and all clusters were not well separated i.e. contamination in the refractory period. I thought the algorithm was merging too much and played around with parameters to increase the number of resulting clusters, because manual merging is easier than splitting.
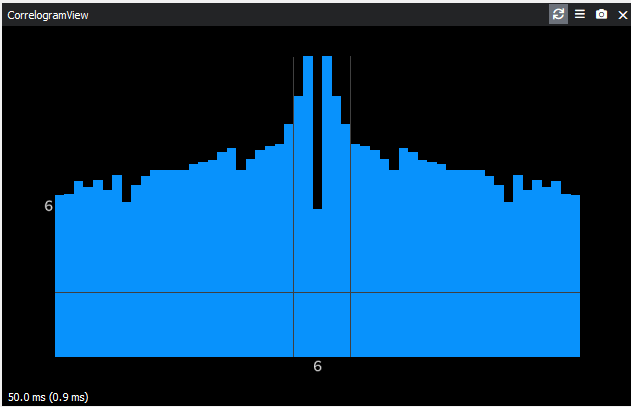
I found that lowering
[clustering] sensitivitythe most dramatically increases the number of clusters. Using[clustering] sensitivity = 0.8gives about 5 or more clusters per channel. However, most clusters still have refractory period violation.In the raw data trace, I can visually tell that there are a few distinct populations in the spike amplitudes. However I noticed that the algorithm puts spikes with apparent difference in amplitudes into one cluster. In some cases, one cluster has two distinct peaks in amplitude:

So far I played around with the following parameters:
[clustering] nb_eltsto 0.95 takes longer time to calculate but does not quite improve the isolation[clustering] cc_mergeto 1 does not quite improve the isolation[clustering] sensitivityto 0.8 or even 0.5 makes calculation time longer, and results in obvious increase of number of clusters[clustering] dispersionto (2, 2) does not quite improve the isolation[detection] N_tto 2 ms has slight improvement in the isolationI would be grateful if I get some advice on how to improve the sorting quality. Thank you very much in advance.
The text was updated successfully, but these errors were encountered: