New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
[BUG 🐛] division by zero error in estimate_foreground_ml #44
Comments
|
Hi @eyaler, thank you for opening this issue! Could you kindly provide code, the image and the alpha matte that yield the division-by-zero-error? That would be very helpful. Thanks! |
|
The only place where I could imagine that division by zero errors could occur is when dividing by the determinant here:
A positive regularization value should in theory ensure that the determinant is positive: https://www.wolframalpha.com/input/?i=%28%28a%5E2+%2B+d%29+*+%28%281+-+a%29%5E2+%2B+d%29+-+%28%281+-+a%29+*+a%29%5E2%29+%3E+0%2C+d+%3E+0 That might not happen when the regularization parameter of Either way, I agree with @tuelwer that we need a minimal reproducible example to debug this. (See test case template: https://github.com/pymatting/pymatting/blob/master/.github/ISSUE_TEMPLATE/bug_report.md) |
|
i was running this with the defaults (reg=1e-5, weight=1). |
|
The alpha mask should be of shape Replace mask=imageio.imread('mask.png')[:,:,:1]/255with mask=imageio.imread('mask.png')[:,:,0].copy()/255and it should work fine. Apparently, Numba simply ignores the shape specified at
I think this issue should be fixed in the Numba library. |
|
i can confirm this solves my issue. until numba fixes it, this could easily be solved by doing
which is just evil... |
|
Sorry about that. This was definitely a mistake in the documentation. Should be fixed now. We could of course do all kinds of checks to see whether the passed arguments are correct and convert them to a more suitable format if necessary, but there is an infinite number of ways to get it wrong, so I do not think that this doable. Just a few examples:
What I am trying to say is, if we start to fix incorrect input, there will be no end to it. The updated documentation (thanks for the tip by the way) will have to do for now. |
|
Type hints might also be a good additional measure to issues like this. Unfortunately, NumPy does not yet support shapes, only I'll leave these issues here so it is easier to follow the development of type hinting for shapes: |
|
We switched back to JIT compilation today because ahead-of-time compilation caused too many issues. Numba's JIT backend is a bit stricter when checking types. |
|
hi, I'm facing the same issue . [ZeroDivisionError: division by zero] trimap function: generating trimap: step 2: now, estimate_alpha_cf is throwing the error of "ZeroDivisionError: division by zero" |
|
I think this is a different issue because the error happens in Anyway, could you create a "minimal, reproducible example" with complete code and images so we can reproduce the issue? |
|
hi @99991 , I am sharing the notebook and corresponding images. |
|
@shamindraparui Thank you, I can reproduce the issue. PyMatting expects that all images are in the range [0, 1], but your img = img / 255.0However, the results are not great because The artifacts can be reduced by using more erosion, but that will remove part of the beak of the bird. It is better to use a few steps of skeletonization in this case, but the result is still not perfect. You might be better off using the original mask you had for that image. What did you use to compute it? import numpy as np
import pymatting
import matplotlib.pyplot as plt
img = pymatting.load_image("blend/ILSVRC2012_test_00000250.jpg", "RGB")
mask = pymatting.load_image("blend/ILSVRC2012_test_00000250.png", "GRAY")
is_fg = (mask > 0.9).astype(np.uint8)
is_bg = (mask < 0.1).astype(np.uint8)
def incorrect_but_good_enough_skeletonization_step(mask):
keep = np.ones(2**9, dtype=np.bool8)
keep[[23, 27, 31, 54, 55, 63, 89, 91, 95, 216, 217, 219, 308, 310, 311,
432, 436, 438, 464, 472, 473, 496, 500, 504]] = 0
padded_mask = np.pad(mask, [(1, 1), (1, 1)], mode="edge")
windows = np.lib.stride_tricks.sliding_window_view(padded_mask, (3, 3))
return mask * keep[windows.reshape(*mask.shape, 9) @ 2**np.arange(9)]
for _ in range(3):
is_fg = incorrect_but_good_enough_skeletonization_step(is_fg)
for _ in range(5):
is_bg = incorrect_but_good_enough_skeletonization_step(is_bg)
trimap = 0.5 + 0.5 * is_fg - 0.5 * is_bg
alpha_cf = pymatting.estimate_alpha_cf(img,trimap,laplacian_kwargs={"epsilon": 1e-2})
alpha_lkm = pymatting.estimate_alpha_lkm(img,trimap,laplacian_kwargs={"radius": 5})
alpha_knn = pymatting.estimate_alpha_knn(img,trimap)
def blend(alpha):
foreground = pymatting.estimate_foreground_ml(img, alpha)
new_background = 0.5 * np.ones_like(foreground)
blended = pymatting.blend(foreground, new_background, alpha)
return blended
composite_cf = blend(alpha_cf)
composite_lkm = blend(alpha_lkm)
composite_knn = blend(alpha_knn)
composite_original = blend(mask)
plt.figure(figsize=(20, 10))
for i, (title, image) in enumerate([
("Alpha CF", alpha_cf),
("Alpha LKM", alpha_lkm),
("Alpha KNN", alpha_knn),
("Original mask", mask),
("Trimap", trimap),
("Composite CF", composite_cf),
("Composite LKM", composite_lkm),
("Composite KNN", composite_knn),
("Composite original", composite_original),
], 1):
plt.subplot(2, 5, i)
plt.title(title)
plt.imshow(image, cmap="gray")
plt.axis("off")
plt.tight_layout()
plt.savefig("plot.png")
plt.show() |
|
@99991 I am trying to compute the salient part of a given image as mask. The algorithm that I am using is U2Net. It is predicting a mask (not so good) which needs to be binarized, trimapped and alpha matted for final overlaying/segmenting the original image. |
|
@shamindraparui Nice, I am looking forward to the results! For animals, there is also https://github.com/JizhiziLi/GFM and it seems to generalize to other prominent image objects to some degree. Maybe it will be helpful for your project. I have added some sanity checks for the input image and trimap. I think this will make it less confusing to provide the input in the expected format. I'll upload it to PyPI later. Until then, it can be installed from source. |
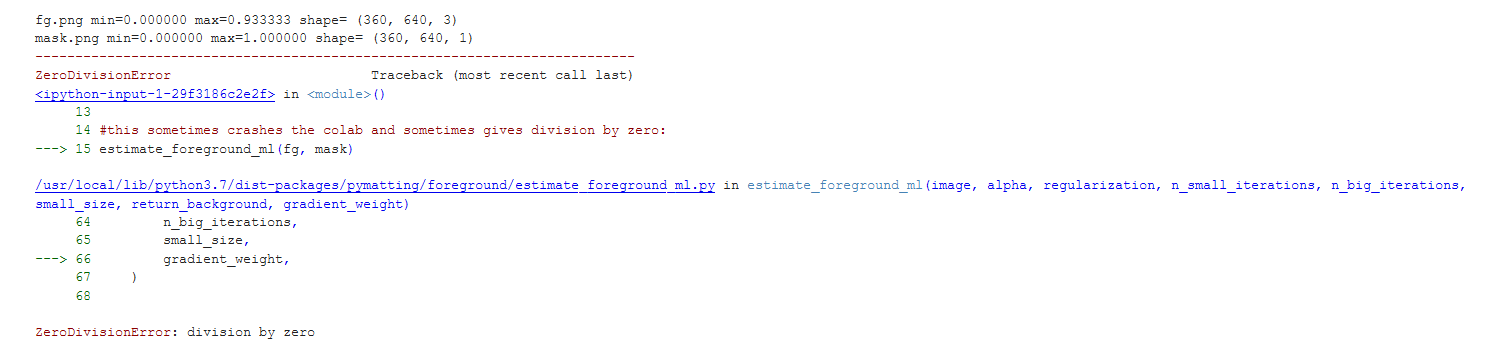

i am getting division by zero errors in estimate_foreground_ml()
what i tried:
the environment is google colab. also sometime this (or something else in pymatting) causes the colab itself to crash and disconnect.
The text was updated successfully, but these errors were encountered: