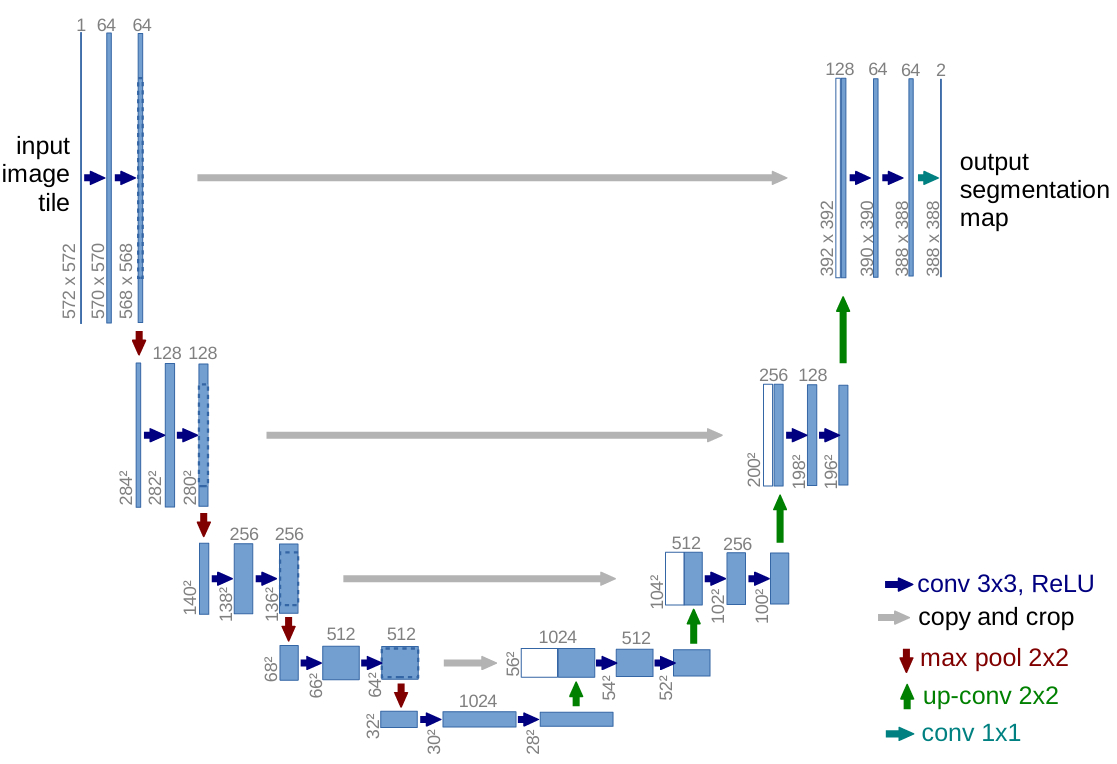
U-Net: Convolutional Networks for Biomedical Image Segmentation
There is large consent that successful training of deep networks requires many thousand annotated training samples. In this paper, we present a network and training strategy that relies on the strong use of data augmentation to use the available annotated samples more efficiently. The architecture consists of a contracting path to capture context and a symmetric expanding path that enables precise localization. We show that such a network can be trained end-to-end from very few images and outperforms the prior best method (a sliding-window convolutional network) on the ISBI challenge for segmentation of neuronal structures in electron microscopic stacks. Using the same network trained on transmitted light microscopy images (phase contrast and DIC) we won the ISBI cell tracking challenge 2015 in these categories by a large margin. Moreover, the network is fast. Segmentation of a 512x512 image takes less than a second on a recent GPU. The full implementation (based on Caffe) and the trained networks are available at this http URL.
| Method | Backbone | Loss | Crop Size | Lr schd | Mem (GB) | Inf time (fps) | Device | mIoU | mIoU(ms+flip) | config | download |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UNet + FCN | UNet-S5-D16 | Cross Entropy | 512x1024 | 160000 | 17.91 | 3.05 | V100 | 69.10 | 71.05 | config | model | log |
| Method | Backbone | Loss | Image Size | Crop Size | Stride | Lr schd | Mem (GB) | Inf time (fps) | Device | mDice | Dice | config | download |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UNet + FCN | UNet-S5-D16 | Cross Entropy | 584x565 | 64x64 | 42x42 | 40000 | 0.680 | - | V100 | 88.38 | 78.67 | config | model | log |
| UNet + FCN | UNet-S5-D16 | Cross Entropy + Dice | 584x565 | 64x64 | 42x42 | 40000 | 0.582 | - | V100 | 88.71 | 79.32 | config | model | log |
| UNet + PSPNet | UNet-S5-D16 | Cross Entropy | 584x565 | 64x64 | 42x42 | 40000 | 0.599 | - | V100 | 88.35 | 78.62 | config | model | log |
| UNet + PSPNet | UNet-S5-D16 | Cross Entropy + Dice | 584x565 | 64x64 | 42x42 | 40000 | 0.585 | - | V100 | 88.76 | 79.42 | config | model | log |
| UNet + DeepLabV3 | UNet-S5-D16 | Cross Entropy | 584x565 | 64x64 | 42x42 | 40000 | 0.596 | - | V100 | 88.38 | 78.69 | config | model | log |
| UNet + DeepLabV3 | UNet-S5-D16 | Cross Entropy + Dice | 584x565 | 64x64 | 42x42 | 40000 | 0.582 | - | V100 | 88.84 | 79.56 | config | model | log |
| Method | Backbone | Loss | Image Size | Crop Size | Stride | Lr schd | Mem (GB) | Inf time (fps) | Device | mDice | Dice | config | download |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UNet + FCN | UNet-S5-D16 | Cross Entropy | 605x700 | 128x128 | 85x85 | 40000 | 0.968 | - | V100 | 89.78 | 81.02 | config | model | log |
| UNet + FCN | UNet-S5-D16 | Cross Entropy + Dice | 605x700 | 128x128 | 85x85 | 40000 | 0.986 | - | V100 | 90.65 | 82.70 | config | model | log |
| UNet + PSPNet | UNet-S5-D16 | Cross Entropy | 605x700 | 128x128 | 85x85 | 40000 | 0.982 | - | V100 | 89.89 | 81.22 | config | model | log |
| UNet + PSPNet | UNet-S5-D16 | Cross Entropy + Dice | 605x700 | 128x128 | 85x85 | 40000 | 1.028 | - | V100 | 90.72 | 82.84 | config | model | log |
| UNet + DeepLabV3 | UNet-S5-D16 | Cross Entropy | 605x700 | 128x128 | 85x85 | 40000 | 0.999 | - | V100 | 89.73 | 80.93 | config | model | log |
| UNet + DeepLabV3 | UNet-S5-D16 | Cross Entropy + Dice | 605x700 | 128x128 | 85x85 | 40000 | 1.010 | - | V100 | 90.65 | 82.71 | config | model | log |
| Method | Backbone | Loss | Image Size | Crop Size | Stride | Lr schd | Mem (GB) | Inf time (fps) | Device | mDice | Dice | config | download |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UNet + FCN | UNet-S5-D16 | Cross Entropy | 960x999 | 128x128 | 85x85 | 40000 | 0.968 | - | V100 | 89.46 | 80.24 | config | model | log |
| UNet + FCN | UNet-S5-D16 | Cross Entropy + Dice | 960x999 | 128x128 | 85x85 | 40000 | 0.986 | - | V100 | 89.52 | 80.40 | config | model | log |
| UNet + PSPNet | UNet-S5-D16 | Cross Entropy | 960x999 | 128x128 | 85x85 | 40000 | 0.982 | - | V100 | 89.52 | 80.36 | config | model | log |
| UNet + PSPNet | UNet-S5-D16 | Cross Entropy + Dice | 960x999 | 128x128 | 85x85 | 40000 | 1.028 | - | V100 | 89.45 | 80.28 | config | model | log |
| UNet + DeepLabV3 | UNet-S5-D16 | Cross Entropy | 960x999 | 128x128 | 85x85 | 40000 | 0.999 | - | V100 | 89.57 | 80.47 | config | model | log |
| UNet + DeepLabV3 | UNet-S5-D16 | Cross Entropy + Dice | 960x999 | 128x128 | 85x85 | 40000 | 1.010 | - | V100 | 89.49 | 80.37 | config | model | log |
| Method | Backbone | Loss | Image Size | Crop Size | Stride | Lr schd | Mem (GB) | Inf time (fps) | Device | mDice | Dice | config | download |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UNet + FCN | UNet-S5-D16 | Cross Entropy | 2336x3504 | 256x256 | 170x170 | 40000 | 2.525 | - | V100 | 88.92 | 79.45 | config | model | log |
| UNet + FCN | UNet-S5-D16 | Cross Entropy + Dice | 2336x3504 | 256x256 | 170x170 | 40000 | 2.623 | - | V100 | 89.64 | 80.87 | config | model | log |
| UNet + PSPNet | UNet-S5-D16 | Cross Entropy | 2336x3504 | 256x256 | 170x170 | 40000 | 2.588 | - | V100 | 89.24 | 80.07 | config | model | log |
| UNet + PSPNet | UNet-S5-D16 | Cross Entropy + Dice | 2336x3504 | 256x256 | 170x170 | 40000 | 2.798 | - | V100 | 89.69 | 80.96 | config | model | log |
| UNet + DeepLabV3 | UNet-S5-D16 | Cross Entropy | 2336x3504 | 256x256 | 170x170 | 40000 | 2.604 | - | V100 | 89.32 | 80.21 | config | model | log |
| UNet + DeepLabV3 | UNet-S5-D16 | Cross Entropy + Dice | 2336x3504 | 256x256 | 170x170 | 40000 | 2.607 | - | V100 | 89.56 | 80.71 | config | model | log |
Note:
- In
DRIVE,STARE,CHASE_DB1, andHRFdataset,mDiceis mean dice of background and vessel, whileDiceis dice metric of vessel(foreground) only.
@inproceedings{ronneberger2015u,
title={U-net: Convolutional networks for biomedical image segmentation},
author={Ronneberger, Olaf and Fischer, Philipp and Brox, Thomas},
booktitle={International Conference on Medical image computing and computer-assisted intervention},
pages={234--241},
year={2015},
organization={Springer}
}