class: middle, center, title-slide count: false
.huge.blue[Matthew Feickert]
.width-02[] matthew.feickert@cern.ch
.width-02[] @HEPfeickert
.width-02[] matthewfeickert
Physics and Astronomy Mini-symposia
[SciPy 2021](https://www.scipy2021.scipy.org/)
July 15th, 2021
.grid[
.kol-1-4.center[
.circle.width-80[ ]
]
CERN
]
.kol-1-4.center[
.circle.width-80[]
University of Illinois
Urbana-Champaign
]
.kol-1-4.center[
.circle.width-80[ ]
]
UCSC SCIPP
]
.kol-1-4.center[
.circle.width-75[ ]
]
National Center for
Supercomputing Applications/Illinois
]
]
.kol-3-4.center.bold[pyhf Core Developers]
.kol-1-4.center.bold.gray[funcX Developer]
.kol-1-2.center[
.width-95[ ]
LHC
]
.kol-1-2.center[
.width-95[
]
LHC
]
.kol-1-2.center[
.width-95[ ]
ATLAS
]
.kol-1-1[
.kol-1-2.center[
.width-45[
]
ATLAS
]
.kol-1-1[
.kol-1-2.center[
.width-45[ ]
]
.kol-1-2.center[
.kol-1-2.center[
.width-100[
]
]
.kol-1-2.center[
.kol-1-2.center[
.width-100[ ]
]
.kol-1-2.center[
.width-85[
]
]
.kol-1-2.center[
.width-85[ ]
]
]
]
]
]
]
]
.kol-1-1[
.kol-1-3.center[
.width-100[ ]
Search for new physics
]
.kol-1-3.center[
]
Search for new physics
]
.kol-1-3.center[
.width-100[ ]
]
Make precision measurements ] .kol-1-3.center[ .width-110[[](https://atlas.web.cern.ch/Atlas/GROUPS/PHYSICS/PAPERS/SUSY-2018-31/)]
Provide constraints on models through setting best limits ] ]
- All require .bold[building statistical models] and .bold[fitting models] to data to perform statistical inference
- Model complexity can be huge for complicated searches
- Problem: Time to fit can be .bold[many hours]
- .blue[
pyhfGoal:] Empower analysts with fast fits and expressive models
.kol-3-4[
- Pure Python implementation of ubiquitous high energy physics (HEP) statistical model specification for multi-bin histogram-based analysis
- Supports .bold[multiple computational backends] and optimizers (defaults of NumPy and SciPy)
- JAX, TensorFlow, and PyTorch backends can leverage hardware acceleration (GPUs, TPUs) and automatic differentiation
- Possible to outperform traditional C++ implementations that are default in HEP
- Ways to learn more:
.kol-1-3.center[
.width-100[ ]
]
.kol-1-3.center[
.width-100[
]
]
.kol-1-3.center[
.width-100[ ]
]
]
.kol-1-3.center[
.width-100[

]
.width-85[
]
.width-85[
]
.width-50[] ]
.kol-1-3[
- Cloud service providers give an excellent Functions as a Service (FaaS) platform that can scale elastically
- Example: Running
pyhfacross 25 worker nodes on Google Cloud Platform- Results being plotted as they are streamed back
- Fit of all signal model hypotheses in analysis takes .bold[3 minutes]!
- Powerful resource, but in (academic) sciences experience is still growing
- "Pay for priority" model
- fast and reliable
- requires funding even with nice support from cloud providers ] .kol-2-3[
.center.width-70[
.kol-1-2[
- HPC facilities are more .bold[commonly available] for use in HEP and provide an opportunity to .bold[efficiently perform statistical inference] of LHC data
- Can pose problems with orchestration and efficient scheduling
- Want to leverage
pyhfhardware accelerated backends at HPC sites for real analysis speedup- Reduce fitting time from hours to minutes
- Idea: Deploy a
pyhfbased .bold[(fitting) Function as a Service] to HPC centers - Example use cases:
- Large scale ensemble fits for statistical combinations
- Large dimensional scans of theory parameter space (e.g. Phenomenological Minimal Supersymmetric Standard Model scans)
- Pseudo-experiment generation ("toys")
]
.kol-1-2[
.center.width-90[
 ]
Model that takes over an hour with traditional C++ framework fit in under 1 minute with pyhf on local GPU
]
]
Model that takes over an hour with traditional C++ framework fit in under 1 minute with pyhf on local GPU
]
.kol-1-2[
- Designed to orchestrate .bold[scientific workloads] across .bold[heterogeneous computing resources] (clusters, clouds, and supercomputers) and task execution providers (HTCondor, Slurm, Torque, and Kubernetes)
- Leverages Parsl parallel scripting library for efficient parallelism and managing concurrent task execution
- Allows users to .bold[register] and then .bold[execute] Python functions in "serverless supercomputing" workflow ] .kol-1-2[
funcXSDK provides a Python API tofuncXservice- Controlling "endpoint" and submission machine can be .bold[totally seperate] (communications routed through Globus)
- Allows for "fire and forget" remote execution where you can run on your laptop, close it, and then retrieve output later
- Tool in a growing ecosystem of distributed computing
- Currently looking into other tools like
Dask.distributedandDask-Jobqueueas well ]
- Currently looking into other tools like
- Deployment of
funcXendpoints is straightforward - Example: Deployment to University of Chicago's RIVER cluster with Kubernetes
.center.width-100[]
.kol-2-5[
- funcX endpoint: logical entity that represents a compute resource
- Managed by an agent process allowing the funcX service to dispatch .bold[user defined functions] to resources for execution
- Agent handles:
- Authentication (Globus) and authorization
- Provisioning of nodes on the compute resource
- Monitoring and management
- Through funcX endpoint config can use expert knowledge of resource to optimize for task ] .kol-3-5[
.tiny[
from funcx_endpoint.endpoint.utils.config import Config
from funcx_endpoint.executors import HighThroughputExecutor
from funcx_endpoint.providers.kubernetes.kube import KubernetesProvider
from funcx_endpoint.strategies import KubeSimpleStrategy
from parsl.addresses import address_by_route
config = Config(
executors=[
HighThroughputExecutor(
max_workers_per_node=1,
address=address_by_route(),
strategy=KubeSimpleStrategy(max_idletime=3600),
container_type="docker",
scheduler_mode="hard",
provider=KubernetesProvider(
init_blocks=0,
min_blocks=1,
max_blocks=100,
init_cpu=2,
max_cpu=3,
init_mem="2000Mi",
max_mem="4600Mi",
image="bengal1/pyhf-funcx:3.8.0.3.0-1",
worker_init="pip freeze",
namespace="servicex",
incluster_config=True,
),
)
],
# ...
)] ]
.kol-2-3[ .tiny[
import json
from time import sleep
import pyhf
from funcx.sdk.client import FuncXClient
from pyhf.contrib.utils import download
def prepare_workspace(data, backend):
import pyhf
pyhf.set_backend(backend)
return pyhf.Workspace(data)
def infer_hypotest(workspace, metadata, patches, backend):
import time
import pyhf
pyhf.set_backend(backend)
tick = time.time()
model = workspace.model(...)
data = workspace.data(model)
test_poi = 1.0
return {
"metadata": metadata,
"cls_obs": float(
pyhf.infer.hypotest(test_poi, data, model, test_stat="qtilde")
),
"fit-time": time.time() - tick,
}
...]
]
.kol-1-3[
- As the analyst user, define the functions that you want the funcX endpoint to execute
- These are run as individual jobs and so require all dependencies of the function to .bold[be defined inside the function]
.tiny[
import numpy # Not in execution scope
def example_function():
import pyhf # Import here
...
pyhf.set_backend("jax") # To use here] ]
.kol-2-3[ .tiny[
...
def main(args):
...
# Initialize funcX client
fxc = FuncXClient()
fxc.max_requests = 200
with open("endpoint_id.txt") as endpoint_file:
pyhf_endpoint = str(endpoint_file.read().rstrip())
# register functions
prepare_func = fxc.register_function(prepare_workspace)
# execute background only workspace
bkgonly_workspace = json.load(bkgonly_json)
prepare_task = fxc.run(
bkgonly_workspace, backend, endpoint_id=pyhf_endpoint, function_id=prepare_func
)
# retrieve function execution output
workspace = None
while not workspace:
try:
workspace = fxc.get_result(prepare_task)
except Exception as excep:
print(f"prepare: {excep}")
sleep(10)
...] ] .kol-1-3[
- With the user functions defined, they can then be registered with the funcX client locally
fx.register_function(...)
- The local funcX client can then execute the request to the remote funcX endpoint, handling all communication and authentication required
fx.run(...)
- While the jobs run on the remote HPC system, can make periodic requests for finished results
fxc.get_result(...)- Returning the output of the user defined functions ]
.kol-2-3[ .tiny[
...
# register functions
infer_func = fxc.register_function(infer_hypotest)
patchset = pyhf.PatchSet(json.load(patchset_json))
# execute patch fits across workers and retrieve them when done
n_patches = len(patchset.patches)
tasks = {}
for patch_idx in range(n_patches):
patch = patchset.patches[patch_idx]
task_id = fxc.run(
workspace,
patch.metadata,
[patch.patch],
backend,
endpoint_id=pyhf_endpoint,
function_id=infer_func,
)
tasks[patch.name] = {"id": task_id, "result": None}
while count_complete(tasks.values()) < n_patches:
for task in tasks.keys():
if not tasks[task]["result"]:
try:
result = fxc.get_result(tasks[task]["id"])
tasks[task]["result"] = result
except Exception as excep:
print(f"inference: {excep}")
sleep(15)
...] ] .kol-1-3[
- The workflow
fx.register_function(...)fx.run(...)
can now be used to scale out .bold[as many custom functions as the workers can handle]
- This allows for all the signal patches (model hypotheses) in a full analysis to be .bold[run simultaneously across HPC workers]
- Run from anywhere (e.g. laptop)!
- The user analyst has .bold[written only simple pure Python]
- No system specific configuration files needed ]
.kol-1-2[
- .bold[Example]: Fitting all 125 models from
pyhfpallet for published ATLAS SUSY 1Lbb analysis - Wall time .bold[under 2 minutes 30 seconds]
- Downloading of
pyhfpallet from HEPData (submit machine) - Registering functions (submit machine)
- Sending serialization to funcX endpoint (remote HPC)
- funcX executing all jobs (remote HPC)
- funcX retrieving finished job output (submit machine)
- Downloading of
- Deployments of funcX endpoints currently used for testing
- University of Chicago River HPC cluster (CPU)
- NCSA Bluewaters (CPU)
- XSEDE Expanse (GPU JAX) ] .kol-1-2[ .tiny[
feickert@ThinkPad-X1:~$ time python fit_analysis.py -c config/1Lbb.json
prepare: waiting-for-ep
prepare: waiting-for-ep
--------------------
<pyhf.workspace.Workspace object at 0x7fb4cfe614f0>
Task C1N2_Wh_hbb_1000_0 complete, there are 1 results now
Task C1N2_Wh_hbb_1000_100 complete, there are 2 results now
Task C1N2_Wh_hbb_1000_150 complete, there are 3 results now
Task C1N2_Wh_hbb_1000_200 complete, there are 4 results now
Task C1N2_Wh_hbb_1000_250 complete, there are 5 results now
Task C1N2_Wh_hbb_1000_300 complete, there are 6 results now
Task C1N2_Wh_hbb_1000_350 complete, there are 7 results now
Task C1N2_Wh_hbb_1000_400 complete, there are 8 results now
Task C1N2_Wh_hbb_1000_50 complete, there are 9 results now
Task C1N2_Wh_hbb_150_0 complete, there are 10 results now
...
Task C1N2_Wh_hbb_900_150 complete, there are 119 results now
Task C1N2_Wh_hbb_900_200 complete, there are 120 results now
inference: waiting-for-ep
Task C1N2_Wh_hbb_900_300 complete, there are 121 results now
Task C1N2_Wh_hbb_900_350 complete, there are 122 results now
Task C1N2_Wh_hbb_900_400 complete, there are 123 results now
Task C1N2_Wh_hbb_900_50 complete, there are 124 results now
Task C1N2_Wh_hbb_900_250 complete, there are 125 results now
--------------------
...
real 2m17.509s
user 0m6.465s
sys 0m1.561s
] ]
.kol-1-2[
- .bold[Example]: Fitting all 125 models from
pyhfpallet for published ATLAS SUSY 1Lbb analysis - Wall time .bold[under 2 minutes 30 seconds]
- Downloading of
pyhfpallet from HEPData (submit machine) - Registering functions (submit machine)
- Sending serialization to funcX endpoint (remote HPC)
- funcX executing all jobs (remote HPC)
- funcX retrieving finished job output (submit machine)
- Downloading of
- Deployments of funcX endpoints currently used for testing
- University of Chicago River HPC cluster (CPU)
- NCSA Bluewaters (CPU)
- XSEDE Expanse (GPU JAX) ] .kol-1-2.center[
.center.width-115[ ]
.center.bold[Click me to watch an asciinema!]
]
]
.center.bold[Click me to watch an asciinema!]
]
.kol-1-2[
- Remember, the returned output is just the .bold[function's return]
- Our hypothesis test user function from earlier:
.tiny[
def infer_hypotest(workspace, metadata, patches, backend):
import time
import pyhf
pyhf.set_backend(backend)
tick = time.time()
model = workspace.model(...)
data = workspace.data(model)
test_poi = 1.0
return {
"metadata": metadata,
"cls_obs": float(
pyhf.infer.hypotest(
test_poi, data, model, test_stat="qtilde"
)
),
"fit-time": time.time() - tick,
}] ] .kol-1-2[
- Allowing for easy and rapid serialization and manipulation of results
- Time from submitting jobs to plot can be minutes
.tiny[
feickert@ThinkPad-X1:~$ python fit_analysis.py -c config/1Lbb.json
# Returned results saved in results.json
feickert@ThinkPad-X1:~$ jq .C1N2_Wh_hbb_1000_0.result results.json
{
"metadata": {
"name": "C1N2_Wh_hbb_1000_0",
"values": [
1000,
0
]
},
"cls_obs": 0.5856783708143126,
"fit-time": 24.032342672348022
}
feickert@ThinkPad-X1:~$ jq .C1N2_Wh_hbb_1000_0.result.cls_obs results.json
0.5856783708143126
] ]
.kol-2-5[
- The nature of FaaS that makes it highly scalable also leads to a problem for taking advantage of just-in-time (JIT) compiled functions
- JIT is super helpful for performing pseudo-experiment generation
- To leverage JITed functions there needs to be .bold[memory that is preserved across invocations] of that function
- FaaS: Each function call is self contained and .bold[doesn't know about global state]
- funcX endpoint listens on a queue and invokes functions
- Still need to know and tune funcX config to specifics of endpoint resource
- No magic bullet when using HPC center batch systems ] .kol-3-5[
.tiny[ ```ipython In [1]: import jax.numpy as jnp ...: from jax import jit, random
In [2]: def selu(x, alpha=1.67, lmbda=1.05): ...: return lmbda * jnp.where(x > 0, x, alpha * jnp.exp(x) - alpha) ...:
In [3]: key = random.PRNGKey(0) ...: x = random.normal(key, (1000000,))
In [4]: %timeit selu(x) 850 µs ± 35.4 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
In [5]: selu_jit = jit(selu)
In [6]: %timeit selu_jit(x) 17.2 µs ± 105 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)
]
.center[50X speedup from JIT]
]
---
# Summary
- Through the combined use of the pure-Python libraries .bold[funcX and `pyhf`], demonstrated the ability to .bold[parallelize and accelerate] statistical inference of physics analyses on HPC systems through a .bold[(fitting) FaaS solution]
- Without having to write any bespoke batch jobs, inference can be registered and executed by analysts with a client Python API that still .bold[achieves the large performance gains] compared to single node execution that is a typical motivation of use of batch systems.
- Allows for transparently switching workflows between .bold[provider systems] and from .bold[CPU to GPU] environments
- Not currently able to leverage benefits of .bold[JITed operations]
- Looking for ways to bridge this
- All [code](https://github.com/matthewfeickert/talk-scipy-2021) used .bold[public and open source] on GitHub!
.kol-1-1[
.kol-1-3.center[
.width-90[[](https://github.com/scikit-hep/pyhf)]
]
.kol-1-3.center[]
.kol-1-3.center[
<br>
.width-100[[](https://github.com/funcx-faas/funcX)]
]
]
---
class: middle
.center[
# Thanks for listening!
# Come talk with us!
.large[[www.scikit-hep.org/pyhf](https://scikit-hep.org/pyhf/)]
]
.grid[
.kol-1-3.center[
.width-90[[](https://scikit-hep.org/)]
]
.kol-1-3.center[
<br>
<!-- .width-90[[](https://github.com/scikit-hep/pyhf)] -->
.width-90[[](https://github.com/scikit-hep/pyhf)]
]
.kol-1-3.center[
<br>
<br>
.width-100[[](https://iris-hep.org/)]
]
]
---
class: end-slide, center
.large[Backup]
---
# funcX endpoints on HPC: Config Example
.kol-1-2[
Example Parsl `HighThroughputExecutor` config (from [Parsl docs](https://parsl.readthedocs.io/en/1.1.0/userguide/execution.html#configuration)) that .bold[funcX extends]
.tiny[
```python
from parsl.config import Config
from libsubmit.providers.local.local import Local
from parsl.executors import HighThroughputExecutor
config = Config(
executors=[
HighThroughputExecutor(
label='local_htex',
workers_per_node=2,
provider=Local(
min_blocks=1,
init_blocks=1,
max_blocks=2,
nodes_per_block=1,
parallelism=0.5
)
)
]
)
] .tiny[
- block: Basic unit of resources acquired from a provider
max_blocks: Maximum number of blocks that can be active per executornodes_per_block: Number of nodes requested per blockparallelism: Ratio of task execution capacity to the sum of running tasks and available tasks ] ] .kol-1-2[ .center.width-100[ ]
]- 9 tasks to compute
- Tasks are allocated to the first block until its
task_capacity(here 4 tasks) reached - Task 5: First block full and
5/9 > parallelism
so Parsl provisions a new block for executing the remaining tasks ]
-
funcX provides a managed service secured by Globus Auth
-
Endpoints can be set up by a site administrator and shared with authorized users through Globus Auth Groups
-
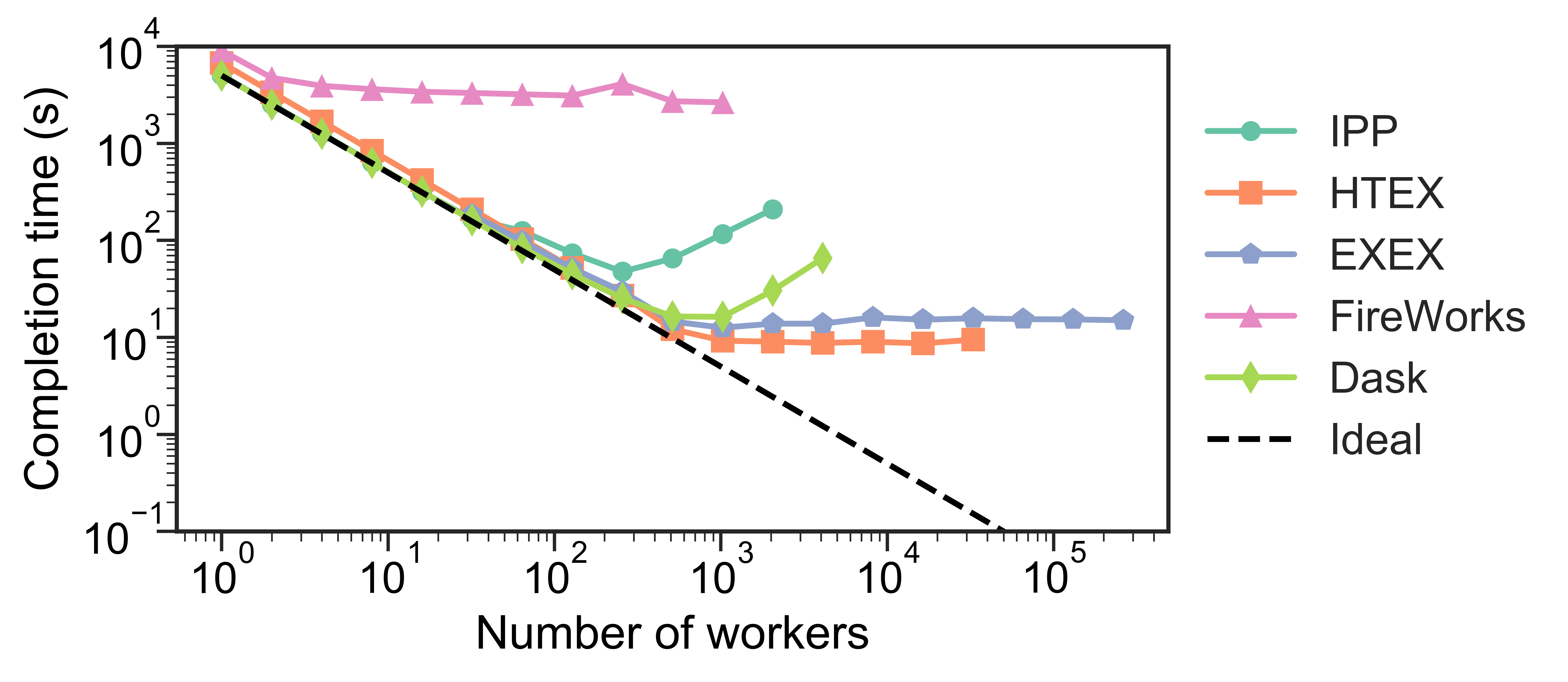
Testing has shown that Dask may not scale well .bold[to thousands of nodes], whereas the funcX High Throughput Executor (HTEX) provided through Parsl scales efficiently
- Lukas Heinrich, .italic[Distributed Gradients for Differentiable Analysis], Future Analysis Systems and Facilities Workshop, 2020.
- Babuji, Y., Woodard, A., Li, Z., Katz, D. S., Clifford, B., Kumar, R., Lacinski, L., Chard, R., Wozniak, J., Foster, I., Wilde, M., and Chard, K., Parsl: Pervasive Parallel Programming in Python. 28th ACM International Symposium on High-Performance Parallel and Distributed Computing (HPDC). 2019. https://doi.org/10.1145/3307681.3325400
class: end-slide, center count: false
The end.


 ]
]