This is official code of MICCAI'2020 PRIME workshop paper:
Uniformizing Techniques to Process CT scans with 3D CNNs for Tuberculosis Prediction (Paper, arXiv)
If you use this code or models in your scientific work, please cite the following paper:
@inproceedings{zunair2020uniformizing,
title={Uniformizing Techniques to Process CT Scans with 3D CNNs for Tuberculosis Prediction},
author={Zunair, Hasib and Rahman, Aimon and Mohammed, Nabeel and Cohen, Joseph Paul},
booktitle={International Workshop on PRedictive Intelligence In MEdicine},
pages={156--168},
year={2020},
organization={Springer}
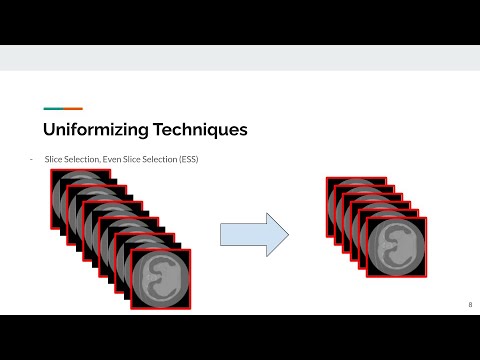
}Data uniformizing methods
- Ubuntu 14.04
- Python 3.6
- Tensorflow: 2.0.0
- Keras: 2.3.1
You can create the appropriate conda environment by running
conda env create -f environment.yml
First, get the data from here. Then:
- Run notebooks in order
others: Contains helper codes to preprocess and visualize samples in dataset.
A 🤗 Spaces demo for detecting pneumonia from CT scans using our method is available here. Demo built by Faizan Shaikh.
More details at this link
Zunair, H., Rahman, A., Mohammed, N.: Estimating Severity from CT Scans
of Tuberculosis Patients using 3D Convolutional Nets and Slice Selection. In:
CLEF2019 Working Notes. Volume 2380 of CEUR Workshop Proceedings.,
Lugano, Switzerland, CEUR-WS.org
<http://ceur-ws.org/Vol-2380>(September 9-12 2019) Previous paper published in CEUR-WS. Paper can be found at CLEF Working Notes 2019 under the section ImageCLEF - Multimedia Retrieval in CLEF.
Your driver's license.