#! sudo apt install bwa/bin/sh: 1: export: : bad variable name
/home/jovyan/.local/bin:/home/jovyan/.local/bin:/srv/conda/bin:/srv/conda/envs/kernel/bin:/srv/npm/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin
!bwaProgram: bwa (alignment via Burrows-Wheeler transformation)
Version: 0.7.17-r1188
Contact: Heng Li <lh3@sanger.ac.uk>
Usage: bwa <command> [options]
Command: index index sequences in the FASTA format
mem BWA-MEM algorithm
fastmap identify super-maximal exact matches
pemerge merge overlapping paired ends (EXPERIMENTAL)
aln gapped/ungapped alignment
samse generate alignment (single ended)
sampe generate alignment (paired ended)
bwasw BWA-SW for long queries
shm manage indices in shared memory
fa2pac convert FASTA to PAC format
pac2bwt generate BWT from PAC
pac2bwtgen alternative algorithm for generating BWT
bwtupdate update .bwt to the new format
bwt2sa generate SA from BWT and Occ
Note: To use BWA, you need to first index the genome with `bwa index'.
There are three alignment algorithms in BWA: `mem', `bwasw', and
`aln/samse/sampe'. If you are not sure which to use, try `bwa mem'
first. Please `man ./bwa.1' for the manual.
test = "abracadabra"
test.find("aca")3
We need index of the source (genome, etc.)
! bwa indexUsage: bwa index [options] <in.fasta>
Options: -a STR BWT construction algorithm: bwtsw, is or rb2 [auto]
-p STR prefix of the index [same as fasta name]
-b INT block size for the bwtsw algorithm (effective with -a bwtsw) [10000000]
-6 index files named as <in.fasta>.64.* instead of <in.fasta>.*
Warning: `-a bwtsw' does not work for short genomes, while `-a is' and
`-a div' do not work not for long genomes.
! bwa index files/NC_005816.fna[bwa_index] Pack FASTA... 0.00 sec
[bwa_index] Construct BWT for the packed sequence...
[bwa_index] 0.00 seconds elapse.
[bwa_index] Update BWT... 0.00 sec
[bwa_index] Pack forward-only FASTA... 0.00 sec
[bwa_index] Construct SA from BWT and Occ... 0.00 sec
[main] Version: 0.7.17-r1188
[main] CMD: bwa index files/NC_005816.fna
[main] Real time: 0.018 sec; CPU: 0.007 sec
! ls files/ACT_example.py ls_orchid.gbk.gz PF05371_seed.faa
alpha.faa make_subsmat.py PF05371_seed.sth
beta.faa m_cold.fasta Plates.csv
clustal_run.py my_blast.xml protein.aln
ec_5.4.2.2.txt my_blat.psl Proux_et_al_2002_Figure_6.py
fasta_dictionary.py my_example.phy resampled.phy
fasta_iterator.py NC_005816.fna simple.dnd
gbvrl1.seq.gz NC_005816.fna.amb single.phy
gbvrl2.seq.gz NC_005816.fna.ann SRR3579118_tiny_1.fastq.gz
getgene.py NC_005816.fna.bwt SRR3579118_tiny_2.fastq.gz
ls_orchid.fasta NC_005816.fna.pac swissprot.py
ls_orchid.gbk NC_005816.fna.sa test.phy
ls_orchid.gbk.bgz NC_005816.gb www_blast.py
ls_orchid.gbk.bz2 opuntia.fasta
! bwa index -p files/yersinia_genome files/NC_005816.fna[bwa_index] Pack FASTA... 0.00 sec
[bwa_index] Construct BWT for the packed sequence...
[bwa_index] 0.00 seconds elapse.
[bwa_index] Update BWT... 0.00 sec
[bwa_index] Pack forward-only FASTA... 0.00 sec
[bwa_index] Construct SA from BWT and Occ... 0.00 sec
[main] Version: 0.7.17-r1188
[main] CMD: bwa index -p files/yersinia_genome files/NC_005816.fna
[main] Real time: 0.019 sec; CPU: 0.007 sec
! ls files/ACT_example.py m_cold.fasta Proux_et_al_2002_Figure_6.py
alpha.faa my_blast.xml resampled.phy
beta.faa my_blat.psl simple.dnd
clustal_run.py my_example.phy single.phy
ec_5.4.2.2.txt NC_005816.fna SRR3579118_tiny_1.fastq.gz
fasta_dictionary.py NC_005816.fna.amb SRR3579118_tiny_2.fastq.gz
fasta_iterator.py NC_005816.fna.ann swissprot.py
gbvrl1.seq.gz NC_005816.fna.bwt test.phy
gbvrl2.seq.gz NC_005816.fna.pac www_blast.py
getgene.py NC_005816.fna.sa yersinia_genome.amb
ls_orchid.fasta NC_005816.gb yersinia_genome.ann
ls_orchid.gbk opuntia.fasta yersinia_genome.bwt
ls_orchid.gbk.bgz PF05371_seed.faa yersinia_genome.pac
ls_orchid.gbk.bz2 PF05371_seed.sth yersinia_genome.sa
ls_orchid.gbk.gz Plates.csv
make_subsmat.py protein.aln
! bwa memUsage: bwa mem [options] <idxbase> <in1.fq> [in2.fq]
Algorithm options:
-t INT number of threads [1]
-k INT minimum seed length [19]
-w INT band width for banded alignment [100]
-d INT off-diagonal X-dropoff [100]
-r FLOAT look for internal seeds inside a seed longer than {-k} * FLOAT [1.5]
-y INT seed occurrence for the 3rd round seeding [20]
-c INT skip seeds with more than INT occurrences [500]
-D FLOAT drop chains shorter than FLOAT fraction of the longest overlapping chain [0.50]
-W INT discard a chain if seeded bases shorter than INT [0]
-m INT perform at most INT rounds of mate rescues for each read [50]
-S skip mate rescue
-P skip pairing; mate rescue performed unless -S also in use
Scoring options:
-A INT score for a sequence match, which scales options -TdBOELU unless overridden [1]
-B INT penalty for a mismatch [4]
-O INT[,INT] gap open penalties for deletions and insertions [6,6]
-E INT[,INT] gap extension penalty; a gap of size k cost '{-O} + {-E}*k' [1,1]
-L INT[,INT] penalty for 5'- and 3'-end clipping [5,5]
-U INT penalty for an unpaired read pair [17]
-x STR read type. Setting -x changes multiple parameters unless overridden [null]
pacbio: -k17 -W40 -r10 -A1 -B1 -O1 -E1 -L0 (PacBio reads to ref)
ont2d: -k14 -W20 -r10 -A1 -B1 -O1 -E1 -L0 (Oxford Nanopore 2D-reads to ref)
intractg: -B9 -O16 -L5 (intra-species contigs to ref)
Input/output options:
-p smart pairing (ignoring in2.fq)
-R STR read group header line such as '@RG\tID:foo\tSM:bar' [null]
-H STR/FILE insert STR to header if it starts with @; or insert lines in FILE [null]
-o FILE sam file to output results to [stdout]
-j treat ALT contigs as part of the primary assembly (i.e. ignore <idxbase>.alt file)
-5 for split alignment, take the alignment with the smallest coordinate as primary
-q don't modify mapQ of supplementary alignments
-K INT process INT input bases in each batch regardless of nThreads (for reproducibility) []
-v INT verbosity level: 1=error, 2=warning, 3=message, 4+=debugging [3]
-T INT minimum score to output [30]
-h INT[,INT] if there are <INT hits with score >80% of the max score, output all in XA [5,200]
-a output all alignments for SE or unpaired PE
-C append FASTA/FASTQ comment to SAM output
-V output the reference FASTA header in the XR tag
-Y use soft clipping for supplementary alignments
-M mark shorter split hits as secondary
-I FLOAT[,FLOAT[,INT[,INT]]]
specify the mean, standard deviation (10% of the mean if absent), max
(4 sigma from the mean if absent) and min of the insert size distribution.
FR orientation only. [inferred]
Note: Please read the man page for detailed description of the command line and options.
! bwa mem files/yersinia_genome files/opuntia.fasta[M::bwa_idx_load_from_disk] read 0 ALT contigs
@SQ SN:gi|45478711|ref|NC_005816.1| LN:9609
@PG ID:bwa PN:bwa VN:0.7.17-r1188 CL:bwa mem files/yersinia_genome files/opuntia.fasta
[M::process] read 7 sequences (6278 bp)...
[M::mem_process_seqs] Processed 7 reads in 0.002 CPU sec, 0.003 real sec
gi|6273291|gb|AF191665.1|AF191665 4 * 0 0 * * 0 0 TATACATTAAAGGAGGGGGATGCGGATAAATGGAAAGGCGAAAGAAAGAAAAAAATGAATCTAAATGATATAGGATTCCACTATGTAAGGTCTTTGAATCATATCATAAAAGACAATGTAATAAAGCATGAATACAGATTCACACATAATTATCTGATATGAATCTATTCATAGAAAAAAGAAAAAAGTAAGAGCCTCCGGCCAATAAAGACTAAGAGGGTTGGCTCAAGAACAAAGTTCATTAAGAGCTCCATTGTAGAATTCAGACCTAATCATTAATCAAGAAGCGATGGGAACGATGTAATCCATGAATACAGAAGATTCAATTGAAAAAGATCCTATGNTCATTGGAAGGATGGCGGAACGAACCAGAGACCAATTCATCTATTCTGAAAAGTGATAAACTAATCCTATAAAACTAAAATAGATATTGAAAGAGTAAATATTCGCCCGCGAAAATTCCTTTTTTATTAAATTGCTCATATTTTCTTTTAGCAATGCAATCTAATAAAATATATCTATACAAAAAAACATAGACAAACTATATATATATATATATATAATATATTTCAAATTCCCTTATATATCCAAATATAAAAATATCTAATAAATTAGATGAATATCAAAGAATCTATTGATTTAGTGTATTATTAAATGTATATATTAATTCAATATTATTATTCTATTCATTTTTATTCATTTTCAAATTTATAATATATTAATCTATATATTAATTTAGAATTCTATTCTAATTCGAATTCAATTTTTAAATATTCATATTCAATTAAAATTGAAATTTTTTCATTCGCGAGGAGCCGGATGAGAAGAAACTCTCATGTCCGGTTCTGTAGTAGAGATGGAATTAAGAAAAAACCATCAACTATAACCCCAAAAGAACCAGA * AS:i:0 XS:i:0
gi|6273290|gb|AF191664.1|AF191664 4 * 0 0 * * 0 0 TATACATTAAAGGAGGGGGATGCGGATAAATGGAAAGGCGAAAGAAAGAAAAAAATGAATCTAAATGATATAGGATTCCACTATGTAAGGTCTTTGAATCATATCATAAAAGACAATGTAATAAAGCATGAATACAGATTCACACATAATTATCTGATATGAATCTATTCATAGAAAAAAGAAAAAAGTAAGAGCCTCCGGCCAATAAAGACTAAGAGGGTTGGCTCAAGAACAAAGTTCATTAAGAGCTCCATTGTAGAATTCAGACCTAATCATTAATCAAGAAGCGATGGGAACGATGTAATCCATGAATACAGAAGATTCAATTGAAAAAGATCCTAATGNTNCATTGGGAAGGATGGCGGAACGAACCAGAGACCAATTCATCTATTCTGAAAAGTGATAAACTAATCCTATAAAACTAAAATAGATATTGAAAGAGTAAATATTCGCCCGCGAAAATTCCTTTTTTATTAAATTGCTCATATTTTCTTTTAGCAATGCAATCTAATAAAATATATCTATACAAAAAAACATAGACAAACTATATATATATATAATATATTTCAAATTCCCTTATATATCCAAATATAAAAATATCTAATAAATTAGATGAATATCAAAGAATCTATTGATTTAGTGTATTATTAAATGTATATATTAATTCAATATTATTATTCTATTCATTTTTATTCATTTTCAAATTTATAATATATTAATCTATATATTAATTTAGAATTCTATTCTAATTCGAATTCAATTTTTAAATATTCATATTCAATTAAAATTGAAATTTTTTCATTCGCGAGGAGCCGGATGAGAAGAAACTCTCATGTCCGGTTCTGTAGTAGAGATGGAATTAAGAAAAAACCATCAACTATAACCCCAAAAGAACCAGA * AS:i:0 XS:i:0
gi|6273289|gb|AF191663.1|AF191663 4 * 0 0 * * 0 0 TATACATTAAAGGAGGGGGATGCGGATAAATGGAAAGGCGAAAGAAAGAAAAAAATGAATCTAAATGATATAGGATTCCACTATGTAAGGTCTTTGAATCATATCATAAAAGACAATGTAATAAAGCATGAATACAGATTCACACATAATTATCTGATATGAATCTATTCATAGAAAAAAGAAAAAAGTAAGAGCCTCCGGCCAATAAAGACTAAGAGGGTTGGCTCAAGAACAAAGTTCATTAAGAGCTCCATTGTAGAATTCAGACCTAATCATTAATCAAGAAGCGATGGGAACGATGTAATCCATGAATACAGAAGATTCAATTGAAAAAGATCCTAATGATTCATTGGGAAGGATGGCGGAACGAACCAGAGACCAATTCATCTATTCTGAAAAGTGATAAACTAATCCTATAAAACTAAAATAGATATTGAAAGAGTAAATATTCGCCCGCGAAAATTCCTTTTTTATTAAATTGCTCATATTTTCTTTTAGCAATGCAATCTAATAAAATATATCTATACAAAAAAACATAGACAAACTATATATATATATAATATATTTCAAATTCCCTTATATATCCAAATATAAAAATATCTAATAAATTAGATGAATATCAAAGAATCTATTGATTTAGTATATTATTAAATGTATATATTAATTCAATATTATTATTCTATTCATTTTTATTCATTTTCAAATTTATAATATATTAATCTATATATTAATTTAGAATTCTATTCTAATTCGAATTCAATTTTTAAATATTCATATTCAATTAAAATTGAAATTTTTTCATTCGCGAGGAGCCGGATGAGAAGAAACTCTCATGTCCGGTTCTGTAGTAGAGATGGAATTAAGAAAAAACCATCAACTATAACCCCAAAAGAACCAGA * AS:i:0 XS:i:0
gi|6273287|gb|AF191661.1|AF191661 4 * 0 0 * * 0 0 TATACATTAAAGAAGGGGGATGCGGATAAATGGAAAGGCGAAAGAAAGAAAAAAATGAATCTAAATGATATACGATTCCACTATGTAAGGTCTTTGAATCATATCATAAAAGACAATGTAATAAAGCATGAATACAGATTCACACATAATTATCTGATATGAATCTATTCATAGAAAAAAGAAAAAAGTAAGAGCCTCCGGCCAATAAAGACTAAGAGGGTTGGCTCAAGAACAAAGTTCATTAAGAGCTCCATTGTAGAATTCAGACCTAATCATTAATCAAGAAGCGATGGGAACGATGTAATCCATGAATACAGAAGATTCAATTGAAAAAGATCCTATGATCCATTGGGAAGGATGGCGGAACGAACCAGAGACCAATTCATCTATTCTGAAAAGTGATAAACTAATCCTATAAAACTAAAATAGATATTGAAAGAGTAAATATTCGCCCGCGAAAATTCCTTTTTTTTTTAAATTGCTCATATTTTATTTTAGCAATGCAATCTAATAAAATATATCTATACAAAAAAATAAAGACAAACTATATATATAATATATTTCAAATTTCCTTATATATCCAAATATAAAAATATCTAATAAATTAGATGAATATCAAAGAATCTATTGATTTAGTGTATTATTAAATGTATATCTTAATTCAATATTATTATTCTATTCATTTTTATTCATTTTCAATTTTATAATATATTAATCTATATATTAATTTATAATTCTATTCTAATTCGAATTCAATTTTTAAATATTCATATTCAATTAAAATTGAAATTTTTTCATTCGCGAGGAGCCGGATGAGAAGAAACTCTCATGTCCGGTTCTGTAGTAGAGATGGAATTAAGAAAAAACCATCAACTATAACCCCAAGAGAACCAGA * AS:i:0 XS:i:0
gi|6273286|gb|AF191660.1|AF191660 4 * 0 0 * * 0 0 TATACATAAAAGAAGGGGGATGCGGATAAATGGAAAGGCGAAAGAAAGAAAAAAATGAATCTAAATGATATACGATTCCACTATGTAAGGTCTTTGAATCATATCATAAAAGACAATGTAATAAAGCATGAATACAGATTCACACATAATTATCTGATATGAATCTATTCATAGAAAAAAGAAAAAAGTAAGAGCCTCCGGCCAATAAAGACTAAGAGGGTTGGCTCAAGAACAAAGTTCATTAAGAGCTCCATTGTAGAATTCAGACCTAATCATTAATCAAGAAGCGATGGGAACGATGTAATCCATGAATACAGAAGATTCAATTGAAAAAGATCCTAATGATCATTGGAAGGATGGCGGAACGAACCAGAGACCAATTCATCTATTCTGAAAAGTGATAAACTAATCCTATAAAACTAAAATAGATATTGAAAGAGTAAATATTCGCCCGCGAAAATTCCTTTTTTATTAAATTGCTCATATTTTATTTTAGCAATGCAATCTAATAAAATATATCTATACAAAAAAATATAGACAAACTATATATATAATATATTTATAATTTCCTTATATATCCAAATATAAAAATATCTAATAAATTAGATGAATATCAAAGAATCTATTGATTTAGTGTATTATTAAATGTATATCTTAATTCAATATTATTATTCTATTCATTTTTATTCATTTTCAAATTTATAATATATTAATCTATATATTAATTTATAATTCTATTCTAATTCGAATTCAATTTTTAAATATTCATATTCAATTAAAATTGAAATTTTTTCATTCGCGAGGAGCCGGATGAGAAGAAACTCTCATGTCCGGTTCTGTAGTAGAGATGGAATTAAGAAAAAACCATCAACTATAACCCCAAGAGAACCAGA * AS:i:0 XS:i:0
gi|6273285|gb|AF191659.1|AF191659 4 * 0 0 * * 0 0 TATACATTAAAGAAGGGGGATGCGGATAAATGGAAAGGCGAAAGAAAGAAAAAAATGAATCTAAATGATATACGATTCCACTATGTAAGGTCTTTGAATCATATCATAAAAGACAATGTAATAAAGCATGAATACAGATTCACACATAATTATCTGATATGAATCTATTCATAGAAAAAAGAAAAAAGTAAGAGCCTCCGGCCAATAAAGACTAAGAGGGTTGGCTCAAGAACAAAGTTCATTAAGAGCTCCATTGTAGAATTCAGACCTAATCATTAATCAAGAAGCGATGGGAACGATGTAATCCATGAATACAGAAGATTCAATTGAAAAAGATCCTAATGATCATTGGGAAGGATGGCGGAACGAACCAGAGACCAATTCATCTATTCTGAAAAGTGATAAACTAATCCTATAAAACTAAAATAGATATTGAAAGAGTAAATATTCGCCCGCGAAAATTCCTTTTTTATTAAATTGCTCATATTTTATTTTAGCAATGCAATCTAATAAAATATATCTATACAAAAAAATATAGACAAACTATATATATAATATATTTCAAATTTCCTTATATACCCAAATATAAAAATATCTAATAAATTAGATGAATATCAAAGAATCCATTGATTTAGTGTATTATTAAATGTATATCTTAATTCAATATTATTATTCTATTCATTTTTATTCATTTTCAAATTTATAATATATTAATCTATATATTAATTTATAATTCTATTCTAATTCGAATTCAATTTTTAAATATTCATATTCAATTAAAATTGAAATTTTTTCATTCGCGAGGAGCCGGATGAGAAGAAACTCTCATGTCCGGTTCTGTAGTAGAGATGGAATTAAGAAAAAACCATCAACTATAACCCCAAGAGAACCAGA * AS:i:0 XS:i:0
gi|6273284|gb|AF191658.1|AF191658 4 * 0 0 * * 0 0 TATACATTAAAGAAGGGGGATGCGGATAAATGGAAAGGCGAAAGAAAGAAAAAAATGAATCTAAATGATATACGATTCCACTATGTAAGGTCTTTGAATCATATCATAAAAGACAATGTAATAAAGCATGAATACAGATTCACACATAATTATCTGATATGAATCTATTCATAGAAAAAAGAAAAAAGTAAGAGCCTCCGGCCAATAAAGACTAAGAGGGTTGGCTCAAGAACAAAGTTCATTAAGAGCTCCATTGTAGAATTCAGACCTAATCATTAATCAAGAAGCGATGGGAACGATGTAATCCATGAATACAGAAGATTCAATTGAAAAAGATCCTAATGATCATTGGGAAGGATGGCGGAACGAACCAGAGACCAATTCATCTATTCTGAAAAGTGATAAACTAATCCTATAAAACTAAAATAGATATTGAAAGAGTAAATATTCGCCCGCGAAAATTCCTTTTTTATTAAATTGCTCATATTTTATTTTAGCAATGCAATCTAATAAAATATATCTATACAAAAAAATATAGACAAACTATATATATATAATATATTTCAAATTTCCTTATATACCCAAATATAAAAATATCTAATAAATTAGATGAATATCAAAGAATCTATTGATTTAGTGTATTATTAAATGTATATCTTAATTCAATATTATTATTCTATTCATTTTTATTCATTTTCAAATTTATAATATATTAATCTATATATTAATTTATAATTCTATTCTAATTCGAATTCAATTTTTAAATATTCATATTCAATTAAAATTGAAATTTTTTCATTCGCGAGGAGCCGGATGAGAAGAAACTCTCATGTCCGGTTCTGTAGTAGAGATGGAATTAAGAAAAAACCATCAACTATAACCCCAAGAGAACCAGA * AS:i:0 XS:i:0
[main] Version: 0.7.17-r1188
[main] CMD: bwa mem files/yersinia_genome files/opuntia.fasta
[main] Real time: 0.004 sec; CPU: 0.004 sec
! bwa mem -o files/opuntia_vs_yersinia.sam files/yersinia_genome files/opuntia.fasta[M::bwa_idx_load_from_disk] read 0 ALT contigs
[M::process] read 7 sequences (6278 bp)...
[M::mem_process_seqs] Processed 7 reads in 0.002 CPU sec, 0.003 real sec
[main] Version: 0.7.17-r1188
[main] CMD: bwa mem -o files/opuntia_vs_yersinia.sam files/yersinia_genome files/opuntia.fasta
[main] Real time: 0.006 sec; CPU: 0.004 sec
! bwa mem -o files/SRR3579118_tiny_1_vs_yersinia.sam \
files/yersinia_genome \
files/SRR3579118_tiny_1.fastq.gz[M::bwa_idx_load_from_disk] read 0 ALT contigs
[M::process] read 100000 sequences (5100000 bp)...
[M::mem_process_seqs] Processed 100000 reads in 2.628 CPU sec, 2.754 real sec
[main] Version: 0.7.17-r1188
[main] CMD: bwa mem -o files/SRR3579118_tiny_1_vs_yersinia.sam files/yersinia_genome files/SRR3579118_tiny_1.fastq.gz
[main] Real time: 3.701 sec; CPU: 3.523 sec
! head -5 files/SRR3579118_tiny_1_vs_yersinia.sam @SQ SN:gi|45478711|ref|NC_005816.1| LN:9609
@PG ID:bwa PN:bwa VN:0.7.17-r1188 CL:bwa mem -o files/SRR3579118_tiny_1_vs_yersinia.sam files/yersinia_genome files/SRR3579118_tiny_1.fastq.gz
SRR3579118.1 4 * 0 0 * * 0 0 NGAGAAAATAACTTTATTTCATTGTGGGGAGCGGGCCGATGTCCAGCCTCA #;<ABGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGEGGGGFFGGGGGGG AS:i:0 XS:i:0
SRR3579118.2 4 * 0 0 * * 0 0 NCCGCTCGCAATCACCCAGATTTCAAGAGCGTGGGTGGCGCCCCGAGAGCC #;<ABECGGGGGGGGGGGGGGGGDFGGGGGGGDGGGGGGGGGGGGGGGGEG AS:i:0 XS:i:0
SRR3579118.3 4 * 0 0 * * 0 0 NCCTGAACCACACTTCAACCTTAAGACCACTGGTGGTGTTATTTCAAAGCC #:<AA@>>>1;=/1;EFEGGGG@FD11CF>@FBB:F9DFGGGGFGEGGGGG AS:i:0 XS:i:0
! samtools Program: samtools (Tools for alignments in the SAM format)
Version: 1.7 (using htslib 1.7-2)
Usage: samtools <command> [options]
Commands:
-- Indexing
dict create a sequence dictionary file
faidx index/extract FASTA
index index alignment
-- Editing
calmd recalculate MD/NM tags and '=' bases
fixmate fix mate information
reheader replace BAM header
targetcut cut fosmid regions (for fosmid pool only)
addreplacerg adds or replaces RG tags
markdup mark duplicates
-- File operations
collate shuffle and group alignments by name
cat concatenate BAMs
merge merge sorted alignments
mpileup multi-way pileup
sort sort alignment file
split splits a file by read group
quickcheck quickly check if SAM/BAM/CRAM file appears intact
fastq converts a BAM to a FASTQ
fasta converts a BAM to a FASTA
-- Statistics
bedcov read depth per BED region
depth compute the depth
flagstat simple stats
idxstats BAM index stats
phase phase heterozygotes
stats generate stats (former bamcheck)
-- Viewing
flags explain BAM flags
tview text alignment viewer
view SAM<->BAM<->CRAM conversion
depad convert padded BAM to unpadded BAM
! samtools flagstat files/SRR3579118_tiny_1_vs_yersinia.sam100000 + 0 in total (QC-passed reads + QC-failed reads)
0 + 0 secondary
0 + 0 supplementary
0 + 0 duplicates
3 + 0 mapped (0.00% : N/A)
0 + 0 paired in sequencing
0 + 0 read1
0 + 0 read2
0 + 0 properly paired (N/A : N/A)
0 + 0 with itself and mate mapped
0 + 0 singletons (N/A : N/A)
0 + 0 with mate mapped to a different chr
0 + 0 with mate mapped to a different chr (mapQ>=5)
! samtools view -F 4 files/SRR3579118_tiny_1_vs_yersinia.samSRR3579118.30391 0 gi|45478711|ref|NC_005816.1| 3612 60 51M * 0 0 AAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTC 3>BBCGGGGGGGGGGGGGGGGGGGEGGGGGGGGGGDGGGGGGGGGGGGGGG NM:i:1 MD:Z:22C28 AS:i:46 XS:i:0
SRR3579118.32159 16 gi|45478711|ref|NC_005816.1| 3658 60 51M * 0 0 TGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGG GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGBCCCB NM:i:1 MD:Z:30C20 AS:i:46 XS:i:0
SRR3579118.54615 16 gi|45478711|ref|NC_005816.1| 3666 60 51M * 0 0 CCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGG AFGGGGGF1GGBGGGGFGGGGGGGGEGGGF@GDGBGDDGGGGGGGAAA:3A NM:i:1 MD:Z:22C28 AS:i:46 XS:i:0
! bwa index -p genome/tiny_human genome/human_tiny.fa.gz[bwa_index] Pack FASTA... 4.52 sec
[bwa_index] Construct BWT for the packed sequence...
[BWTIncCreate] textLength=323945236, availableWord=34793804
[BWTIncConstructFromPacked] 10 iterations done. 57394212 characters processed.
[BWTIncConstructFromPacked] 20 iterations done. 106031796 characters processed.
[BWTIncConstructFromPacked] 30 iterations done. 149256980 characters processed.
[BWTIncConstructFromPacked] 40 iterations done. 187671588 characters processed.
[BWTIncConstructFromPacked] 50 iterations done. 221810580 characters processed.
[BWTIncConstructFromPacked] 60 iterations done. 252149380 characters processed.
[BWTIncConstructFromPacked] 70 iterations done. 279110564 characters processed.
[BWTIncConstructFromPacked] 80 iterations done. 303069700 characters processed.
[BWTIncConstructFromPacked] 90 iterations done. 323945236 characters processed.
[bwt_gen] Finished constructing BWT in 90 iterations.
[bwa_index] 173.72 seconds elapse.
[bwa_index] Update BWT... 4.67 sec
[bwa_index] Pack forward-only FASTA... 3.61 sec
[bwa_index] Construct SA from BWT and Occ... 107.29 sec
[main] Version: 0.7.17-r1188
[main] CMD: bwa index -p genome/tiny_human genome/human_tiny.fa.gz
[main] Real time: 306.027 sec; CPU: 293.818 sec
! zcat files/SRR3579118_tiny_1.fastq.gz | head@SRR3579118.1 HWI:1:X:4:1101:1126:1837/1
NGAGAAAATAACTTTATTTCATTGTGGGGAGCGGGCCGATGTCCAGCCTCA
+
#;<ABGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGEGGGGFFGGGGGGG
@SRR3579118.2 HWI:1:X:4:1101:1250:1851/1
NCCGCTCGCAATCACCCAGATTTCAAGAGCGTGGGTGGCGCCCCGAGAGCC
+
#;<ABECGGGGGGGGGGGGGGGGDFGGGGGGGDGGGGGGGGGGGGGGGGEG
@SRR3579118.3 HWI:1:X:4:1101:1180:1867/1
NCCTGAACCACACTTCAACCTTAAGACCACTGGTGGTGTTATTTCAAAGCC
gzip: stdout: Broken pipe
! zcat files/SRR3579118_tiny_2.fastq.gz | head@SRR3579118.1 HWI:1:X:4:1101:1126:1837/2
ACAAGAACATGTCTGTACACCTGTCCCCCTGCTTCAGGGACGTCCAGATCG
+
CCCCCGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGFGGGGGFG
@SRR3579118.2 HWI:1:X:4:1101:1250:1851/2
CATCTTACGCTGGGACCCCGCCAAGGAGCCCCAGGAAGTAGGTGAAAGGGC
+
CCCBBGGGGGGGGGGGGGGGGGGGGGGGGGGCGFGCFGGGGGGGGGGGGGG
@SRR3579118.3 HWI:1:X:4:1101:1180:1867/2
TCCAATTAAAGTAACACTGGCAACTTTGAAAATGTCTGTACAGCCAACGGT
gzip: stdout: Broken pipe
Warning: do not leave spaces after backslash characters
%%bash
bwa mem \
-o human_srr_test.sam \
genome/tiny_human \
files/SRR3579118_tiny_1.fastq.gz \
files/SRR3579118_tiny_2.fastq.gz[M::bwa_idx_load_from_disk] read 0 ALT contigs
[M::process] read 196080 sequences (10000080 bp)...
[M::process] read 3920 sequences (199920 bp)...
[M::mem_pestat] # candidate unique pairs for (FF, FR, RF, RR): (2, 5510, 9, 0)
[M::mem_pestat] skip orientation FF as there are not enough pairs
[M::mem_pestat] analyzing insert size distribution for orientation FR...
[M::mem_pestat] (25, 50, 75) percentile: (124, 156, 220)
[M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 412)
[M::mem_pestat] mean and std.dev: (157.97, 54.68)
[M::mem_pestat] low and high boundaries for proper pairs: (1, 508)
[M::mem_pestat] skip orientation RF as there are not enough pairs
[M::mem_pestat] skip orientation RR as there are not enough pairs
[M::mem_process_seqs] Processed 196080 reads in 35.699 CPU sec, 36.360 real sec
[M::mem_pestat] # candidate unique pairs for (FF, FR, RF, RR): (0, 96, 0, 0)
[M::mem_pestat] skip orientation FF as there are not enough pairs
[M::mem_pestat] analyzing insert size distribution for orientation FR...
[M::mem_pestat] (25, 50, 75) percentile: (124, 149, 219)
[M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 409)
[M::mem_pestat] mean and std.dev: (156.19, 57.53)
[M::mem_pestat] low and high boundaries for proper pairs: (1, 504)
[M::mem_pestat] skip orientation RF as there are not enough pairs
[M::mem_pestat] skip orientation RR as there are not enough pairs
[M::mem_process_seqs] Processed 3920 reads in 1.283 CPU sec, 1.293 real sec
[main] Version: 0.7.17-r1188
[main] CMD: bwa mem -o human_srr_test.sam genome/tiny_human files/SRR3579118_tiny_1.fastq.gz files/SRR3579118_tiny_2.fastq.gz
[main] Real time: 41.145 sec; CPU: 39.844 sec
! samtools flagstat human_srr_test.sam200000 + 0 in total (QC-passed reads + QC-failed reads)
0 + 0 secondary
0 + 0 supplementary
0 + 0 duplicates
69409 + 0 mapped (34.70% : N/A)
200000 + 0 paired in sequencing
100000 + 0 read1
100000 + 0 read2
53874 + 0 properly paired (26.94% : N/A)
65952 + 0 with itself and mate mapped
3457 + 0 singletons (1.73% : N/A)
5558 + 0 with mate mapped to a different chr
1938 + 0 with mate mapped to a different chr (mapQ>=5)
! head human_srr_test.sam@SQ SN:chr20 LN:64444167
@SQ SN:chr21 LN:46709983
@SQ SN:chr22 LN:50818468
@PG ID:bwa PN:bwa VN:0.7.17-r1188 CL:bwa mem -o human_srr_test.sam genome/tiny_human files/SRR3579118_tiny_1.fastq.gz files/SRR3579118_tiny_2.fastq.gz
SRR3579118.1 77 * 0 0 * * 0 0 NGAGAAAATAACTTTATTTCATTGTGGGGAGCGGGCCGATGTCCAGCCTCA #;<ABGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGEGGGGFFGGGGGGG AS:i:0 XS:i:0
SRR3579118.1 141 * 0 0 * * 0 0 ACAAGAACATGTCTGTACACCTGTCCCCCTGCTTCAGGGACGTCCAGATCG CCCCCGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGFGGGGGFG AS:i:0 XS:i:0
SRR3579118.2 77 * 0 0 * * 0 0 NCCGCTCGCAATCACCCAGATTTCAAGAGCGTGGGTGGCGCCCCGAGAGCC #;<ABECGGGGGGGGGGGGGGGGDFGGGGGGGDGGGGGGGGGGGGGGGGEG AS:i:0 XS:i:0
SRR3579118.2 141 * 0 0 * * 0 0 CATCTTACGCTGGGACCCCGCCAAGGAGCCCCAGGAAGTAGGTGAAAGGGC CCCBBGGGGGGGGGGGGGGGGGGGGGGGGGGCGFGCFGGGGGGGGGGGGGG AS:i:0 XS:i:0
SRR3579118.3 99 chr20 38978103 54 1S50M = 38978167 111 NCCTGAACCACACTTCAACCTTAAGACCACTGGTGGTGTTATTTCAAAGCC #:<AA@>>>1;=/1;EFEGGGG@FD11CF>@FBB:F9DFGGGGFGEGGGGG NM:i:5 MD:Z:5G12T0G9C6A13 MC:Z:4S47M AS:i:25 XS:i:0
SRR3579118.3 147 chr20 38978167 60 4S47M = 38978103 -111 ACCGTTGGCTGTACAGACATTTTCAAAGTTGCCAGTGTTACTTTAATTGGA <C9:FEGFCGGGF1CBFEBF<1GBDC>FE1;;1CEFGEGFF>=1F>B@BAB NM:i:0 MD:Z:47 MC:Z:1S50M AS:i:47 XS:i:0
ASCII characters are used for representing phred scores between 0-40. Version1, add 33 to phred score, Version2 add 64 to phred score
Typical quality distribution for Illumina sequencing
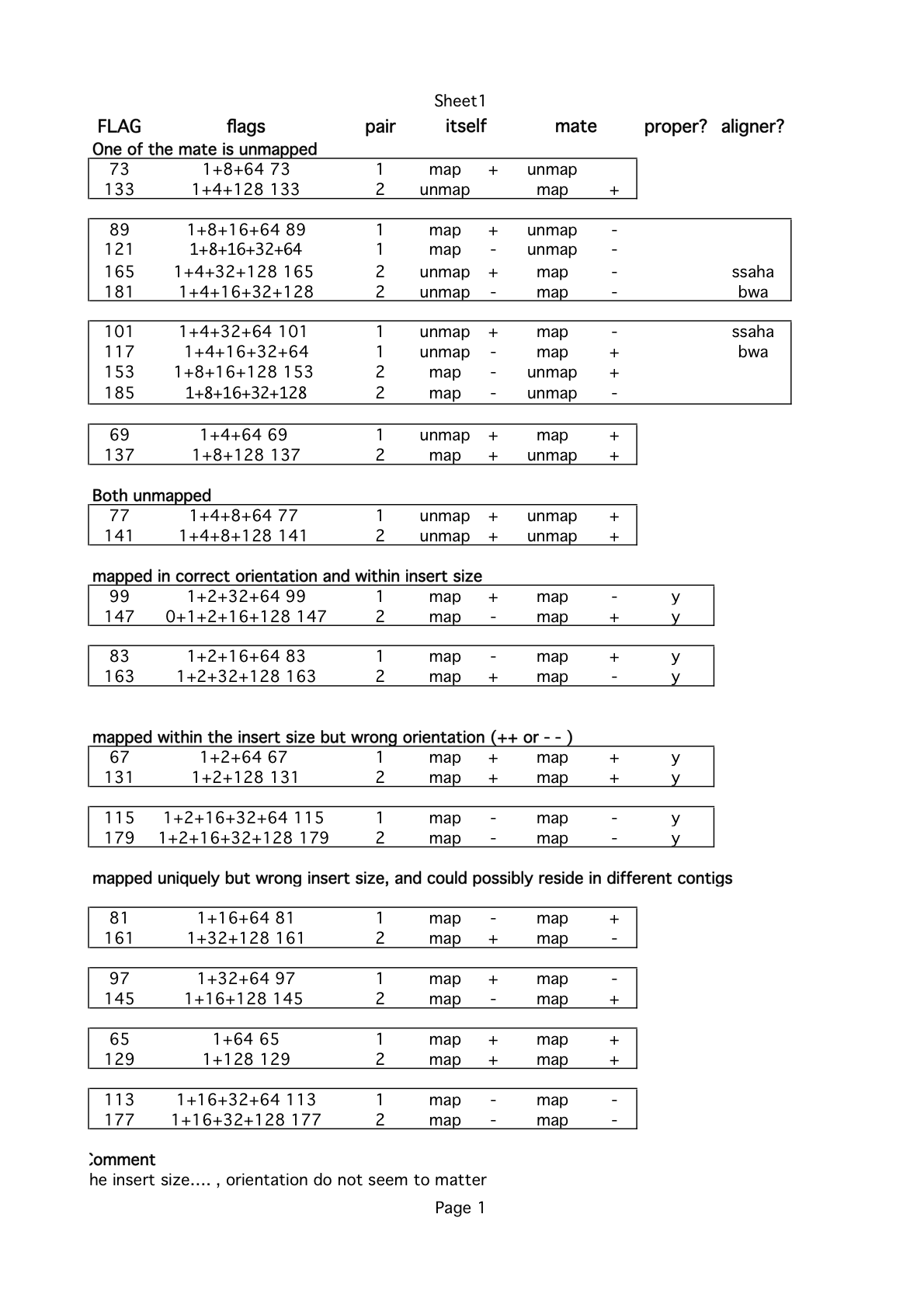
samtools allows filtering reads based on flags (second column). -F means exclude, -f means include. For a list of flags refer to image below
The table below summarizes various conditions as groups
An interactive site can be used to interpret all possible flag values
! samtools view -F 4 human_srr_test.sam | headSRR3579118.3 99 chr20 38978103 54 1S50M = 38978167 111 NCCTGAACCACACTTCAACCTTAAGACCACTGGTGGTGTTATTTCAAAGCC #:<AA@>>>1;=/1;EFEGGGG@FD11CF>@FBB:F9DFGGGGFGEGGGGG NM:i:5 MD:Z:5G12T0G9C6A13 MC:Z:4S47M AS:i:25 XS:i:0
SRR3579118.3 147 chr20 38978167 60 4S47M = 38978103 -111 ACCGTTGGCTGTACAGACATTTTCAAAGTTGCCAGTGTTACTTTAATTGGA <C9:FEGFCGGGF1CBFEBF<1GBDC>FE1;;1CEFGEGFF>=1F>B@BAB NM:i:0 MD:Z:47 MC:Z:1S50M AS:i:47 XS:i:0
SRR3579118.5 185 chr22 42565179 47 51M = 42565179 0 GTCGTCCTCTTCGACCGAGCGCGCAGCTTCGGGAGGGACGCACATGGAGCG <GGGGGGGGFGGGGGGDCBG>C1GGGGGGEBGGE/GGGGGGGBGEGBCBBB NM:i:3 MD:Z:19T2A3T24 AS:i:36 XS:i:0
SRR3579118.9 83 chr20 49722183 0 13S37M1S = 49722053 -167 GTGCTGATCAGTAGTGGGATCGCGCCTGTGAATAGCCACTGCACTCCAGCG FFGGGGGGGGGGGGGGGGGGGGGGGGFGGGGGGGGGGEGGGGGBGCBA?:3 NM:i:2 MD:Z:7T18C10 MC:Z:16S29M6S AS:i:27 XS:i:31
SRR3579118.9 163 chr20 49722053 0 16S29M6S = 49722183 167 GTGCGCTATGCCGATCGGGTGTCCGCACTAAGTTCGGCATCAATATGGTGA CCBBCGGGGGFGGGGGGGGGGGGEGGGGGEEFGF=FGG@GFFGEGGGGGGG NM:i:1 MD:Z:7T21 MC:Z:13S37M1S AS:i:24 XS:i:36 XA:Z:chr20,-50557842,51M,4;chr20,-18258713,51M,6;chr22,-23261942,26M25S,1;
SRR3579118.12 99 chr22 23132744 7 2S36M13S = 23132836 134 NCGCTATGTTGCTCAGGCTGGAGTGCAGTGGCTATTCACAGGCGCGATCCC #;<ABGGGGGGGGGGGGGEGGGGGGGGG1FGBGGGGGGGGFGGGGGG@GBG NM:i:1 MD:Z:29T6 MC:Z:7S42M2S AS:i:31 XS:i:35
SRR3579118.12 147 chr22 23132836 7 7S42M2S = 23132744 -134 TTCCGACCTGGGCCGGTTCACCCCTCCTTAGGCAACCTGGTGGTCCCCCGC GGGGGGGGGGGGGGGGGGGGGGGGGGGGCGGGGGGGGGGGGGGGGGCCCCB NM:i:2 MD:Z:6T21T13 MC:Z:2S36M13S AS:i:32 XS:i:35 XA:Z:chr20,+3094364,3S35M13S,0;chr20,-18258666,14S34M3S,1;chr20,-39349193,51M,5;chr20,-50557781,49M2S,4;chr20,+39352904,7S29M15S,0;
SRR3579118.14 83 chr21 23390752 60 6S45M = 23390673 -124 ATCGCCGTTCTGGTAAAAAGCTGGAAGATGGCCCTAAATTCTTGAAGTCTN #GGGGGGGGGGGFGGGGGGGGGGGGGDCGGGGGGFGGGGFGGGGGFBA=3# NM:i:2 MD:Z:24A19G0 MC:Z:5S46M AS:i:39 XS:i:19
SRR3579118.14 163 chr21 23390673 60 5S46M = 23390752 124 GCGCCGGCTATGCCCCTGTATTGGATTGCCACACGGCTCACATTGCATGCA CCCCBGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG: NM:i:3 MD:Z:14G0C13A16 MC:Z:6S45M AS:i:31 XS:i:0
SRR3579118.15 185 chr22 42565106 18 51M = 42565106 0 TGGAGGTTCTAGCAGGGGAGCGCAGCTACTCGTATACCCTTGACCGAAGAC GGGGGFGGGGGGGGGGGFGGGGGGGGGGGGGGGGGGGGGGGGGGEGCBBCB NM:i:0 MD:Z:51 AS:i:51 XS:i:45 XA:Z:chr20,-18295226,51M,2;
samtools view: writing to standard output failed: Broken pipe
samtools view: error closing standard output: -1
! samtools view -F 4 human_srr_test.sam | wc -l69409
! samtools view -f 4 human_srr_test.sam | wc -l130591
! samtools view -F 99 human_srr_test.sam | wc -l0
! samtools view -F 2 human_srr_test.sam | wc -l146126
! samtools view -f 99 -f 147 human_srr_test.sam | wc -l0
Due to weird path environment issues, we need to run the following code every time.. We should find better alternative..
import os
existing_path = os.environ['PATH']
os.environ['PATH']= existing_path + ':/home/jovyan/bin/gatk-4.1.0.0/'! gatk Usage template for all tools (uses --spark-runner LOCAL when used with a Spark tool)
gatk AnyTool toolArgs
Usage template for Spark tools (will NOT work on non-Spark tools)
gatk SparkTool toolArgs [ -- --spark-runner <LOCAL | SPARK | GCS> sparkArgs ]
Getting help
gatk --list Print the list of available tools
gatk Tool --help Print help on a particular tool
Configuration File Specification
--gatk-config-file PATH/TO/GATK/PROPERTIES/FILE
gatk forwards commands to GATK and adds some sugar for submitting spark jobs
--spark-runner <target> controls how spark tools are run
valid targets are:
LOCAL: run using the in-memory spark runner
SPARK: run using spark-submit on an existing cluster
--spark-master must be specified
--spark-submit-command may be specified to control the Spark submit command
arguments to spark-submit may optionally be specified after --
GCS: run using Google cloud dataproc
commands after the -- will be passed to dataproc
--cluster <your-cluster> must be specified after the --
spark properties and some common spark-submit parameters will be translated
to dataproc equivalents
--dry-run may be specified to output the generated command line without running it
--java-options 'OPTION1[ OPTION2=Y ... ]' optional - pass the given string of options to the
java JVM at runtime.
Java options MUST be passed inside a single string with space-separated values.