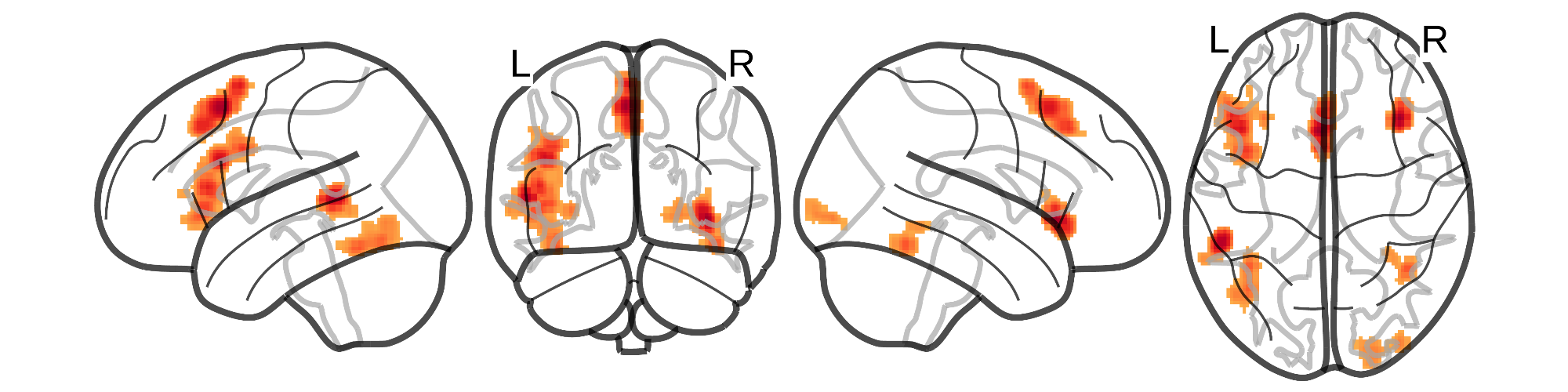
This repository contains the code, data, and results for the following paper:
@article{enge2021,
title = {A meta-analysis of {{fMRI}} studies of semantic cognition in children},
author = {Enge, Alexander and Abdel Rahman, Rasha and Skeide, Michael A.},
year = {2021},
journal = {NeuroImage},
volume = {241},
pages = {118436},
issn = {1053-8119},
doi = {https://doi.org/10.1016/j.neuroimage.2021.118436}
}
In the respective folders, you can view the data that went into the meta-analysis, the Python code that was used to perform it, and the results (statistical maps, tables, and figures).
If you want rerun or modify any of the code, you have three different options.
Simply hit the "launch Binder" badge at the top or this link to open an interactive version of the repository on a cloud server (kindly provided by the Binder project). Please note that launching the server may take a couple of minutes and that the computational resources (CPU cores and memory) are limited.
If you want to run the code on your local computer, we suggest you do so using our Docker container. This will create a small, Linux-based virtual machine which already has all software packages installed. To do so, first download and install Docker Desktop. Once Docker Desktop is running, open a command line window (called "Terminal" on Linux/Mac or "PowerShell" on Windows). From there, execute the following:
docker run --rm -p 8888:8888 skeidelab/meta_semantics:v1.1
You will see a couple of URLs, the last one of which you need to copy and paste into the search bar of your web browser. From there, you will be able to access, execute, and modify our Python notebooks interactively.
Note that Singularity is an open source alterantive to Docker which will also allow you to run the container – even on systems where you do not have root user priveleges.
We recommend using one of the two containerized solutions above because they will ensure that you are executing the code in the exact same software environment which we have also used for our paper. However, you can also execute the code directly on your local system.
To do so, please follow these three steps:
-
Make sure you have a recent version (> 3.6) of Python installed. You can test this by typing
python3 --versionon the command line. If you do not yet have Python installed, you can get it e.g. as a part of the Anaconda toolkit. -
Clone the repository from GitHub, either by (a) downloading it as a zip file or by (b) opening the command line and typing
git pull https://github.com/SkeideLab/meta_semantics.git. Note that the latter requires you to have a local installation of git. -
Install the necessary Python packages by opening a new command line, navigating into the directory that you have just downloaded (e.g.,
cd meta_semantics), and executing the following command:pip3 install -U -r requirements.txt.
Then you can use your IDE of choice (e.g., the Spyder IDE shipped with Anaconda) to open and run our Python scripts locally.
We are glad to receive any feedback, questions, or criticisms on this project. Simply open an issue on GitHub or use the corresponding author's e-mail address as provided in the paper.
Thanks a lot for your time and interest.