-
Notifications
You must be signed in to change notification settings - Fork 88
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Running simulation on HPCC #150
Comments
|
We would definitely need more information about your situation to even begin to try to help you debug it. From your error, I'm guessing your .svg files have been corrupted - either they weren't completely written/saved or there's a memory error or something else. You'd want to be sure you haven't run up against limits on your HPC account, like storage limits for your output files or runtime limits, although you claim that your simulation continues to run. A few suggestions/questions:
|
|
cancer_immune.zip
|
|
The version I'm using is 1.9.1. I didn't try the original cancer immune 3D on HPCC. What you have is the same as the ones I have in the beginning. The error message shows up after about 30 images (or even later). |
|
For starters, I'd strongly recommend updating to the latest PhysiCell release. I created a repo where I did that, using your code and
That repo is at https://github.com/rheiland/tony_model . I can let this model run longer on my laptop at some point to see if I get the mangled .svg you reported. But it would be good if you could try it on your HPC system too. Out of curiosity, is there some reason you are running a 3D model, as opposed to a 2D version? I'm just thinking the initial debugging and results would go smoother. Then switch to 3D once you're confident in the 2D results? |
|
Just pushed an updated main.cpp that uses the _v2 version of saving full data. |
|
I will try to use the latest version on HPCC and see if that makes any difference. As for the model, our project requires to build a 3D instead of a 2D model, so I chose to start with 3D model. For the cells' positions, I used the given data to assign positions for every cancer cell initially. In this case, red cells denote the cancer cells while grey cells denote healthy cells (tissue). After a certain period of time, I introduced immune cells (colored in purple) in the middle as a spheroid. By the way, I believe the parameters that I had in this setting are incorrect. In general, every cell should be adjacent to each other in the beginning instead of splitting up into several neighborhoods. I should have used a smaller number for cell to cell adhesion strength and bigger number for cell to cell repulsion strength. I will talk to the HPCC tech support again with your updates, and I will follow up if the problem is still unsolved. |
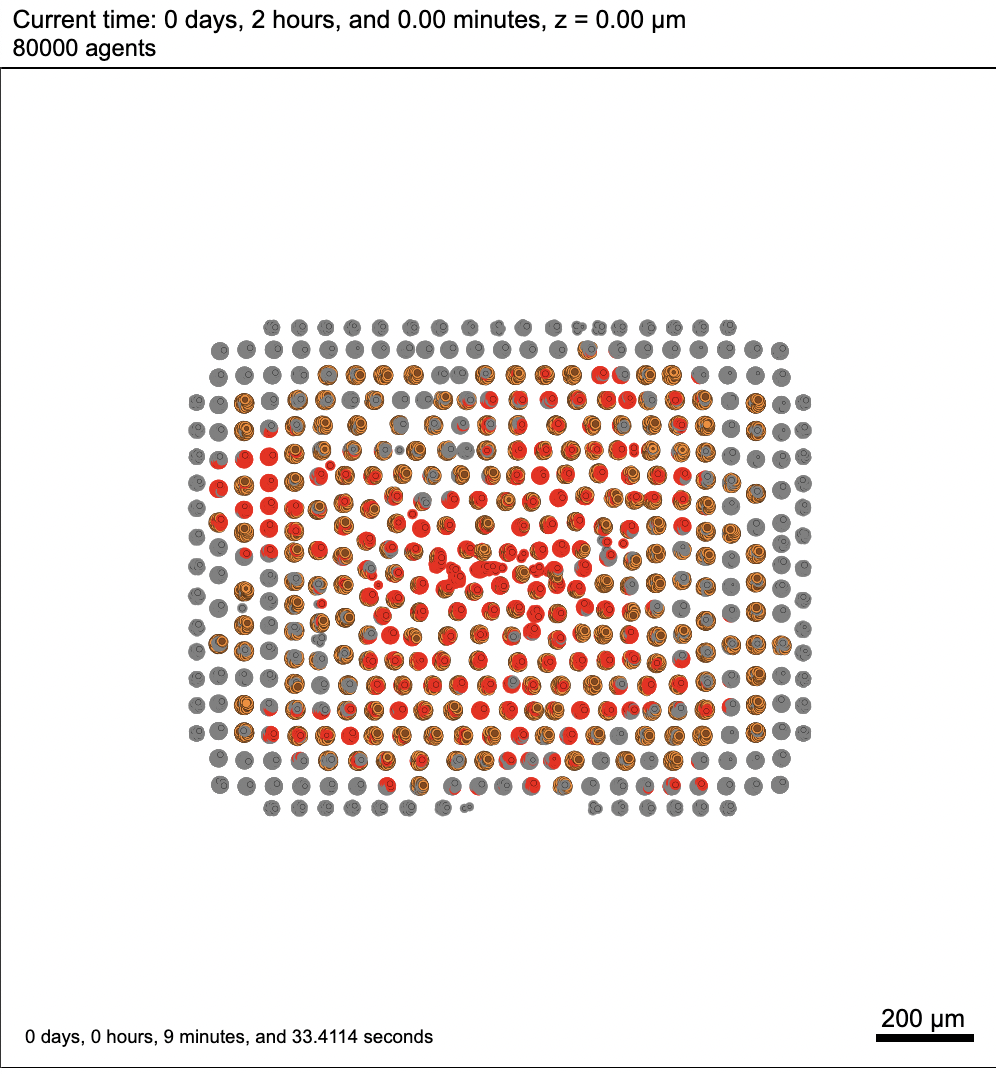
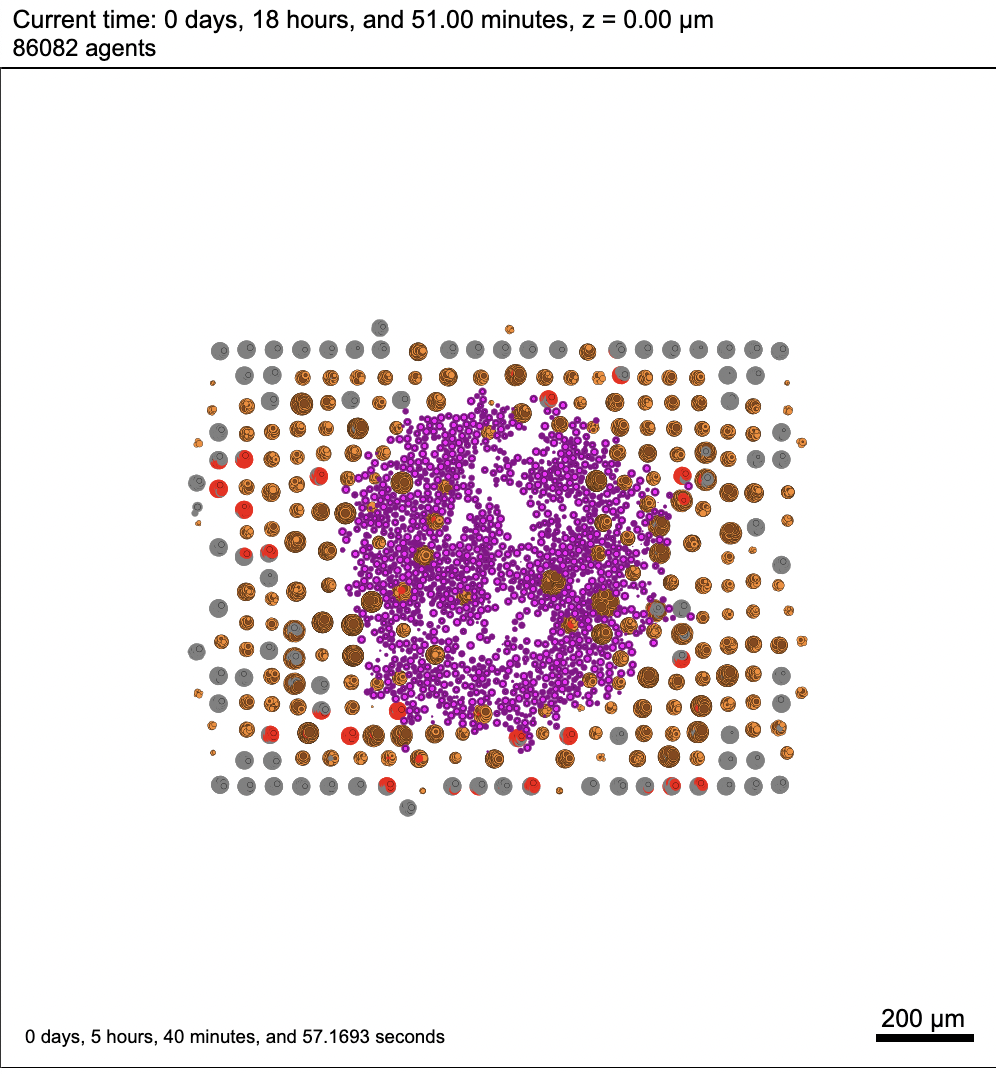

I've run a few simulations on my laptop and everything works fine. Now I am trying to use the HPCC at my university to test the simulation, but I encountered this error while viewing the output images.

I have no idea what this error means exactly. I also notice that for the first few minutes of the simulation, there is nothing wrong with the images, however, this error message comes up in the middle of the simulation after a while. In the meantime, this error doesn't stop my simulation, instead, it keeps running till the end but the rest of the images are all having the same issue.
The text was updated successfully, but these errors were encountered: