New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
speeding up the transform command
#92
Comments
|
Just recording a suggestion from @RossDeVito for us to use multi-processing! from functools import partial
import numpy as np
from multiprocessing import Pool
def single_check(idxs, arr):
return np.all(arr[:, idxs], axis=1)
def parallel_loop(idxs, arr, num_haps, n_processes=None):
with Pool(processes=n_processes) as pool:
res = pool.map(partial(single_check, arr=arr), idxs)
return np.stack(res, axis=1)I was initially hesitant to use this because the In any case, I would still like to explore other options to improve the speed first. I know that we'll ultimately have to use multi-processing in the end, but I'd like to leave it as the method of last resort, especially since other strategies we implement may change everything anyway? This code will definitely be helpful to have for then. |
|
Another idea from Tara is to use |
|
Another thing to consider is that we could load the data from Also, we would first need to check whether it's possible to do all of our operations on packed bit arrays or chunks of data at a time. If not, then we could consider writing our own compiled Cython extension for parts of this command, but that's a whole thing. |
|
@Ayimany and I discussed this issue today, and we've begun to identify a path forward! We can probably implement multiple strategies, all of which should speed things up by a bit
|
|
I've used numba to great success with variable length data (inspired by all of Awkward Array being built with numba). For example, I use it in GenVarLoader to construct haplotypes with indels about fast as numpy can copy reference sequence into an output array (i.e. import numpy as np
import numba as nb
# enable parallel execution
@nb.njit(nogil=True, parallel=True, cache=True)
def _transform(
arr: npt.NDArray[np.bool_],
idxs: npt.NDArray[np.integer],
idx_offsets: npt.NDArray[np.integer],
out: npt.NDArray[np.bool_]
):
# use nb.prange() to mark loops that are parallelizable
for i in nb.prange(len(idx_offsets) - 1):
hap_idx = idxs[idx_offsets[i] : idx_offsets[i+1]]
for _j in nb.prange(len(hap_idx)):
j = hap_idx[_j]
out[:, i] &= arr[:, j]I’d also recommend speed profiling with py-spy to confirm that this is the bottleneck if you haven't already. (Can also recommend memray for memory profiling.) |
|
Thank you so much, @d-laub ! I'm looking forward to trying this!! |

Description
Transforming a set of haplotypes can take a while, especially if you have many haplotypes. A quick benchmark via the
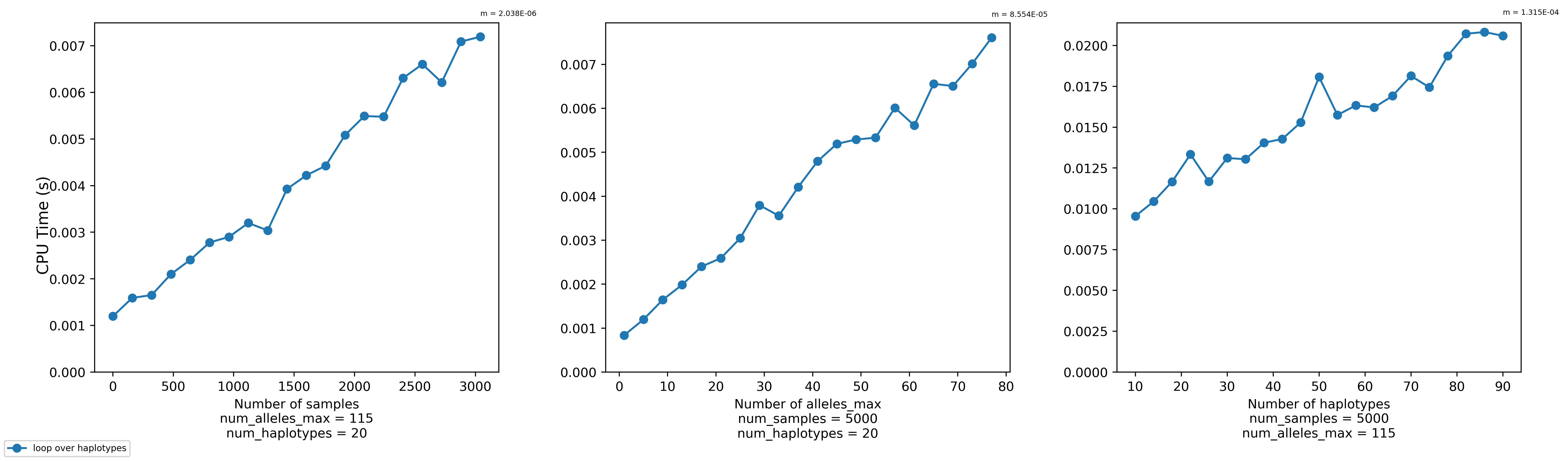
bench_transform.pyscript seems to indicate that it scales linearly with the number of samples, the max number of alleles in each haplotype, and the number of haplotypes.@s041629 wants to transform roughly 21,000 haplotypes for 500,000 samples where the max number of alleles in a haplotype is 115. Currently (as of f45742f), the
Haplotypes.transform()function takes about 10-25 real minutes to accomplish this task. (It varies based on the willingness of the kernel to prioritize our job on the node.)For now, I think this will be fast enough. But we might want to brainstorm some ideas for the future. In any case, I don't really think I can justify spending a lot more time to improve this right now (since I've already spent quite a bit), so I'm gonna take a break and solicit suggestions until I feel energized enough to tackle this again.
Details
The vast majority of the time seems to be spent in this for-loop.
haptools/haptools/data/haplotypes.py
Lines 1054 to 1055 in 639985e
Unfortunately, I can't use broadcasting to speed up this loop because the size of
idxs[i]might vary on each iteration. And I can't pad the array to make broadcasting work because I quickly run out of memory for arrays of that shape. Ideally, I would like to find a solution that doesn't require multi-threading or the installation of new dependencies besides those we have already.Steps to reproduce
A basic test
This test uses just 5 samples, 4 variants, and 3 haplotypes. It's a sanity check to make sure the
Haplotypes.transform()function works and that none of the code will fail.A benchmarking test
To benchmark the function and see how it scales with the number of samples, max number of alleles in each haplotype, and the number of haplotypes in each allele, you can use the
bench_transform.pyscript as so. It will create a plot like the one I've included above.haptools/tests/bench_transform.py
Lines 19 to 21 in 639985e
A pure numpy-based test of the for-loop
The tests above require that you install the dev setup for
haptools. If you'd like to reproduce just the problematic for-loop for @s041629's situation in pure numpy you can do something like this:A full, long-running test
If requested, I can provide files that reproduce the entire setup so that you can execute the
transformcommand with them. The files are a bit too large to attach here.Some things I've already tried
arr(akaequality_arr) of size equal to the number of haplotypes and then performingnp.all()on the other dimension. This takes longer and requires too much more memory. See fac79b0.num_alleles < num_haps, at least in this situation). This was a clever strategy from Ryan Eveloff. Strangely, this also took longer than looping over the haplotypes. I'm not sure why. See 335e5f7.Potential ideas
.hapfile Matteo gave me indicates that there would be at least 1,000 leaves in such a tree.The text was updated successfully, but these errors were encountered: